What you'll take away
Genome integrity characterization via linkage analysis with digital PCR means months-long assay development and complicated bioinformatics.
See how Countable PCR overcomes digital PCR limitations — so your genome integrity characterization is
- Drastically faster — go from assay optimization to linkage insights in 3 days, not months
- More straightforward — get precise transgene counts without manual thresholding
- Much simpler — get automated linkage information without complex bioinformatics
Video transcript
I'm Kyle Kim, RA here at Countable Labs, and I want to share with you today a really cool DNA linkage analysis assay for cell and gene therapy applications using Countable PCR.
The accurate characterization of complete versus partial transgene packaging in viral vectors is super critical for the efficacy and safety of gene therapy products, and it's often one of the biggest challenges in the cell and gene therapy space.
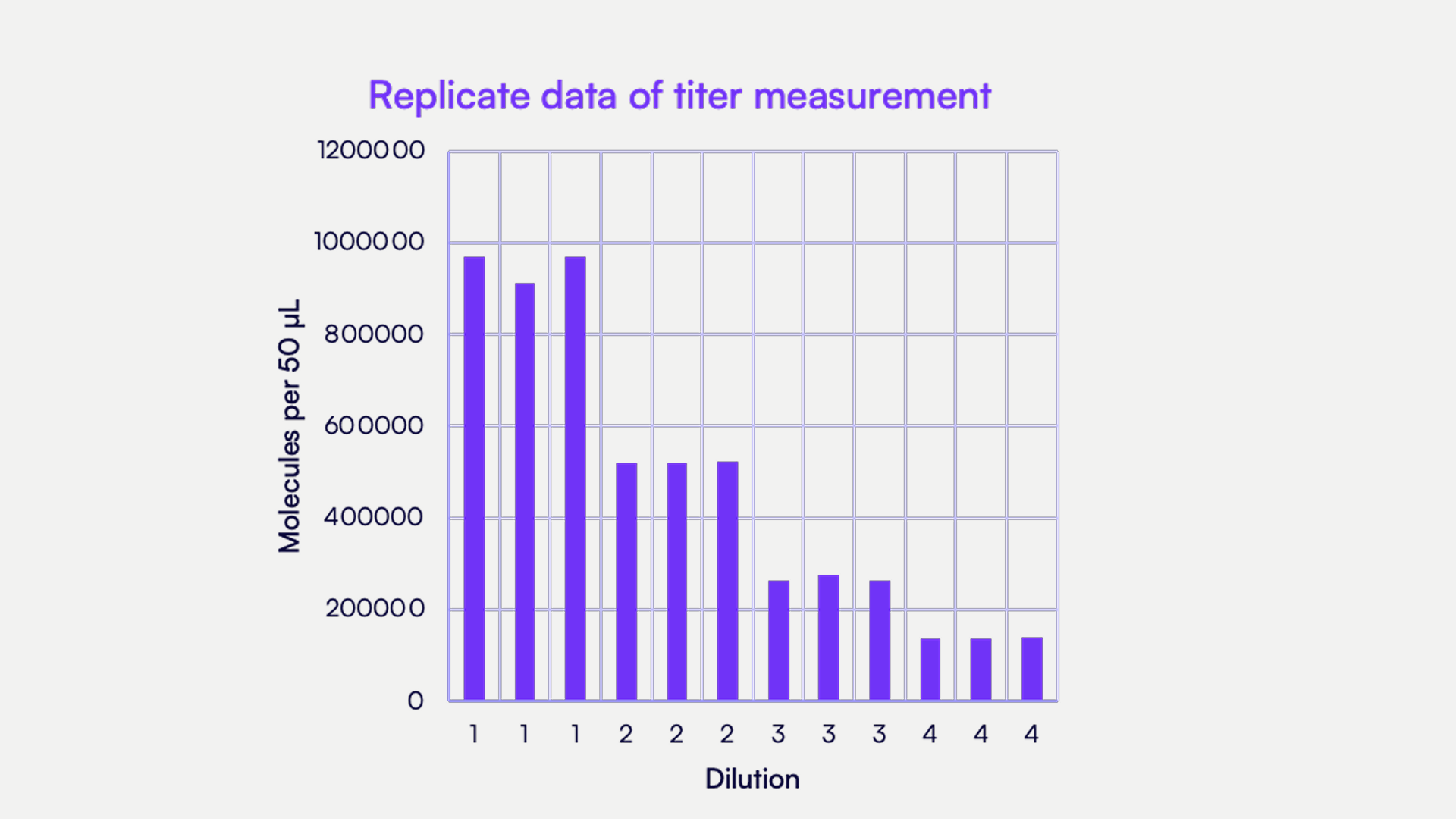
Countable PCR makes DNA linkage assays more straightforward than in digital PCR due to two key features. The first is single-molecule isolation in a 3D matrix, which enables you to get counts with unmatched precision. The second is the use of our Universal Multiplexing probes, which allows you to design and optimize your assay in a matter of days rather than the weeks or months it may take with digital PCR and TaqMan probes.
Let me show you an example in which I targeted three regions of a transgene. In this experiment, I targeted the CMV region, a GFP reporter gene, and a poly-A tail of a vector plasmid. Universal Multiplexing uses adapters on standard PCR primers to tag target amplicons with your probes, making it more affordable and faster to iterate than with TaqMan probes.
Countable Labs' single-molecule isolation in a 3D matrix with over 30 million compartments also makes characterizing complete versus partial transgene packaging highly accurate. This 3D matrix ensures that each molecule is isolated in a single compartment. Amplification of that compartment provides information on what target or targets may be present in that single molecule. If two or more targets are present in that molecule, they would be considered linked.
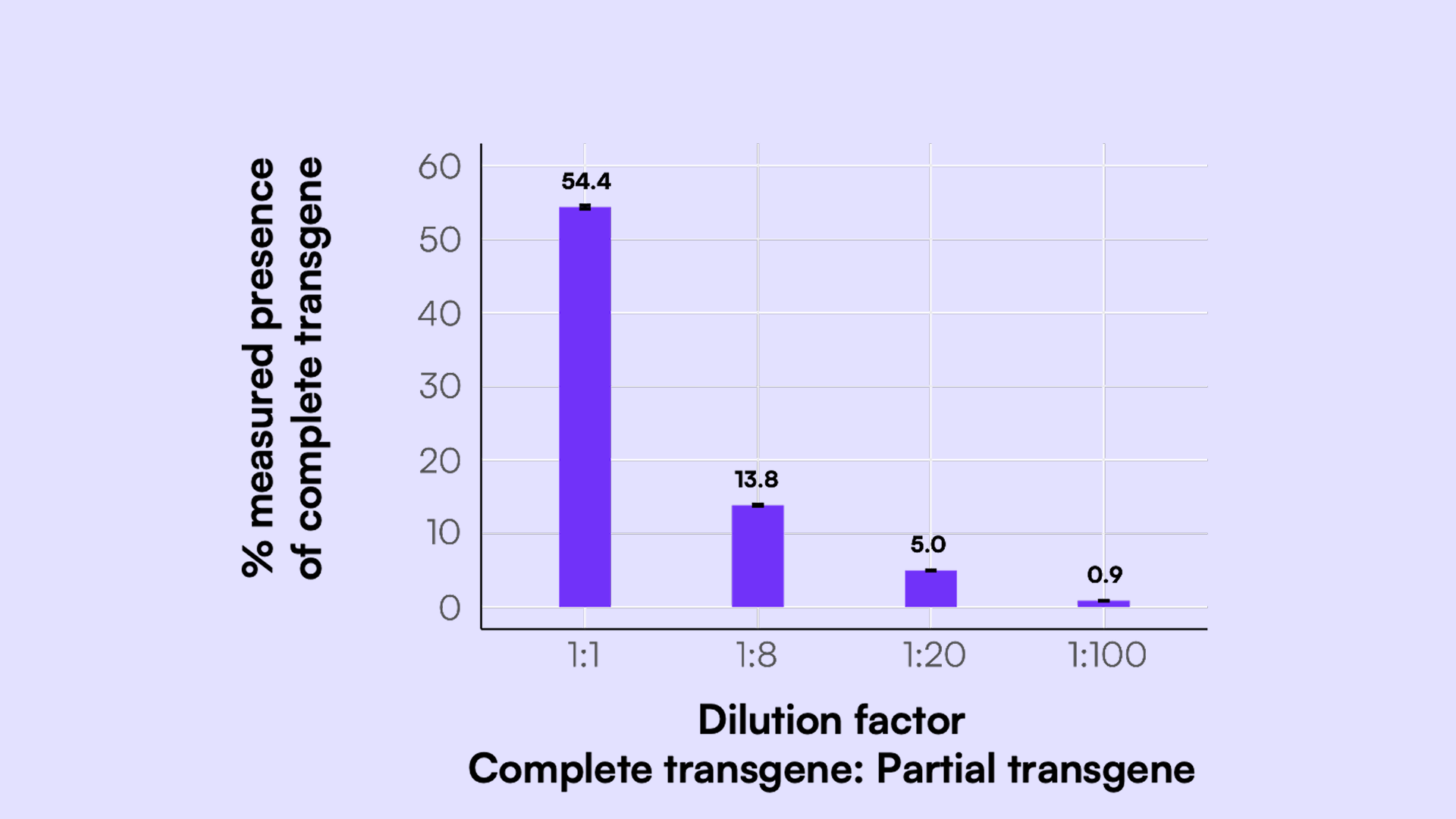
In this scheme, channels 1, 2, and 3—blue, green, and orange—represent a complete transgene. In instances where the transgene is not fully packaged, I would expect a mixture of partial signals. This is an output of our Countable PCR system, in which the three different light sheets represent the three different targets of our assay. The system automatically overlays the images, quantifies, and runs the DNA linkage analysis.
When only one or two targets are present in the compartment, you see a partial signal. For example, an orange compartment contains GFP and a poly-A portion. A blue compartment indicates that the molecule only has the CMV enhancer. White compartments, where all three signals are overlaid, represent complete transgenes with all three components.
To summarize: from generating the initial primer design to receiving the final data, the whole process took me only about three days. Our universal multiplexing kit makes it super easy to interrogate multiple targets using only standard PCR primers to target your regions of interest.
Additionally, because our system performs the counting and linkage analysis automatically, there’s no need for elaborate bioinformatics or manual thresholding. At the end of the run, you receive an output for the counts for each channel, as well as information on what fraction they co-occupy.
If you're interested in running DNA linkage analysis for easy multiplexing experiments, please feel free to reach out to one of our specialists at Countable Labs today. Thank you.
What you'll take away
Genome integrity characterization via linkage analysis with digital PCR means months-long assay development and complicated bioinformatics.
See how Countable PCR overcomes digital PCR limitations — so your genome integrity characterization is
- Drastically faster — go from assay optimization to linkage insights in 3 days, not months
- More straightforward — get precise transgene counts without manual thresholding
- Much simpler — get automated linkage information without complex bioinformatics
Video transcript
I'm Kyle Kim, RA here at Countable Labs, and I want to share with you today a really cool DNA linkage analysis assay for cell and gene therapy applications using Countable PCR.
The accurate characterization of complete versus partial transgene packaging in viral vectors is super critical for the efficacy and safety of gene therapy products, and it's often one of the biggest challenges in the cell and gene therapy space.
Countable PCR makes DNA linkage assays more straightforward than in digital PCR due to two key features. The first is single-molecule isolation in a 3D matrix, which enables you to get counts with unmatched precision. The second is the use of our Universal Multiplexing probes, which allows you to design and optimize your assay in a matter of days rather than the weeks or months it may take with digital PCR and TaqMan probes.
Let me show you an example in which I targeted three regions of a transgene. In this experiment, I targeted the CMV region, a GFP reporter gene, and a poly-A tail of a vector plasmid. Universal Multiplexing uses adapters on standard PCR primers to tag target amplicons with your probes, making it more affordable and faster to iterate than with TaqMan probes.
Countable Labs' single-molecule isolation in a 3D matrix with over 30 million compartments also makes characterizing complete versus partial transgene packaging highly accurate. This 3D matrix ensures that each molecule is isolated in a single compartment. Amplification of that compartment provides information on what target or targets may be present in that single molecule. If two or more targets are present in that molecule, they would be considered linked.
In this scheme, channels 1, 2, and 3—blue, green, and orange—represent a complete transgene. In instances where the transgene is not fully packaged, I would expect a mixture of partial signals. This is an output of our Countable PCR system, in which the three different light sheets represent the three different targets of our assay. The system automatically overlays the images, quantifies, and runs the DNA linkage analysis.
When only one or two targets are present in the compartment, you see a partial signal. For example, an orange compartment contains GFP and a poly-A portion. A blue compartment indicates that the molecule only has the CMV enhancer. White compartments, where all three signals are overlaid, represent complete transgenes with all three components.
To summarize: from generating the initial primer design to receiving the final data, the whole process took me only about three days. Our universal multiplexing kit makes it super easy to interrogate multiple targets using only standard PCR primers to target your regions of interest.
Additionally, because our system performs the counting and linkage analysis automatically, there’s no need for elaborate bioinformatics or manual thresholding. At the end of the run, you receive an output for the counts for each channel, as well as information on what fraction they co-occupy.
If you're interested in running DNA linkage analysis for easy multiplexing experiments, please feel free to reach out to one of our specialists at Countable Labs today. Thank you.


.png)