High-resolution and precise genome integrity in three days – no bioinformatics needed
.png)
.png)
When genome integrity is on the line, time and clarity are everything.
If you’re using digital PCR (dPCR) to confirm vector integrity, you’ve likely hit a wall: multi-target linkage analysis takes weeks. Worse, it’s powered by assumptions — statistical models like Poisson distribution and Monte Carlo simulations — that introduce ambiguity.
Countable PCR changes all of that. It delivers high-resolution linkage analysis in just three days – no bioinformatics, no mathematical gymnastics, no guessing.
With dPCR, confirming genome integrity requires estimating co-localization using complex statistical formulas and occupancy models. The software essentially guesses how many partitions might contain a fully linked sequence, versus fragments that happened to land in the same well.
This means the result isn’t what’s observed — it’s what’s inferred.
Countable PCR is different. Using a structured gel-like matrix with ~30 million compartments, it physically separates single molecules before amplification. After PCR, the system simply counts which compartments contain all the regions you’re looking for — no threshold tuning, no λ (lambda) estimates, no Poisson corrections.
You see the actual signal – no math or assumptions required.
With Countable’s Universal Multiplexing Kit, you amplify multiple targets using standard primers with universal adapters. That means:
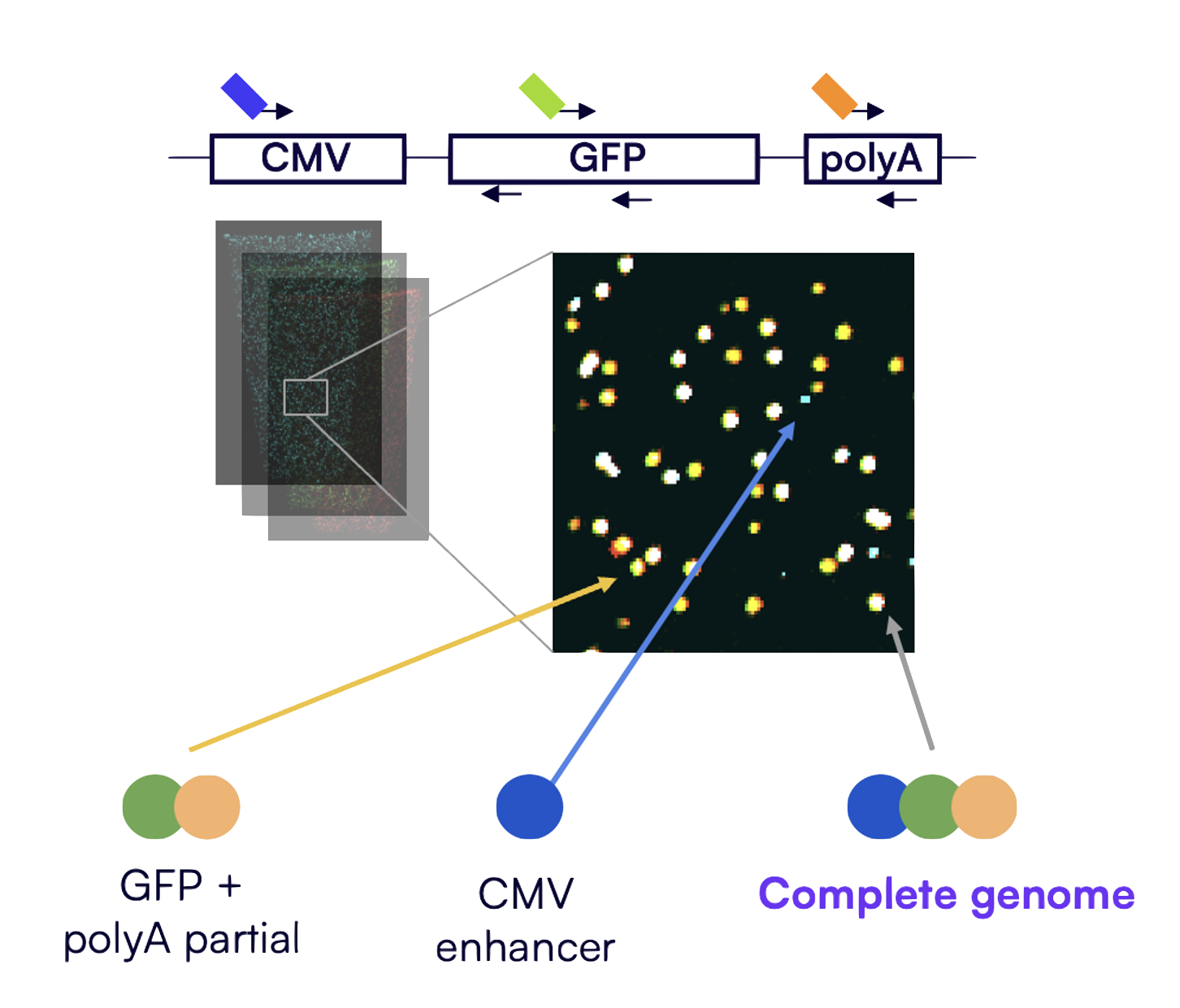
In one case study, researchers targeted three plasmid regions — CMV enhancer, GFP, and poly-A. The Countable System generated composite images from three channels and reported exact counts of full-length and partial sequences — seen in the figure above. A white signal is all three regions — intact and linked. Two colors or fewer means partial genome.
The best part, it took just 3 days from designing this experiment and ordering primers to seeing the results.
Simple, direct, and visual.