Multiplexing with pre-optimized probes – get unbiased amplification and balanced signal out of the box

New to Universal Multiplexing? Start with this blog post before diving in.
There are so many great reasons for performing multiplex PCR experiments. Running more targets per sample lets you reduce consumables usage. It decreases errors from pipetting different samples. And overall, it’s just a better reflection of biology.
But multiplexing is also hard. It means hours in the lab, struggling to get your signal optimized. You tweak and test your probes and primers, only to have assays fail or give you inconclusive results. Repeatedly.
The source of many of these woes with multiplexing is probe design.
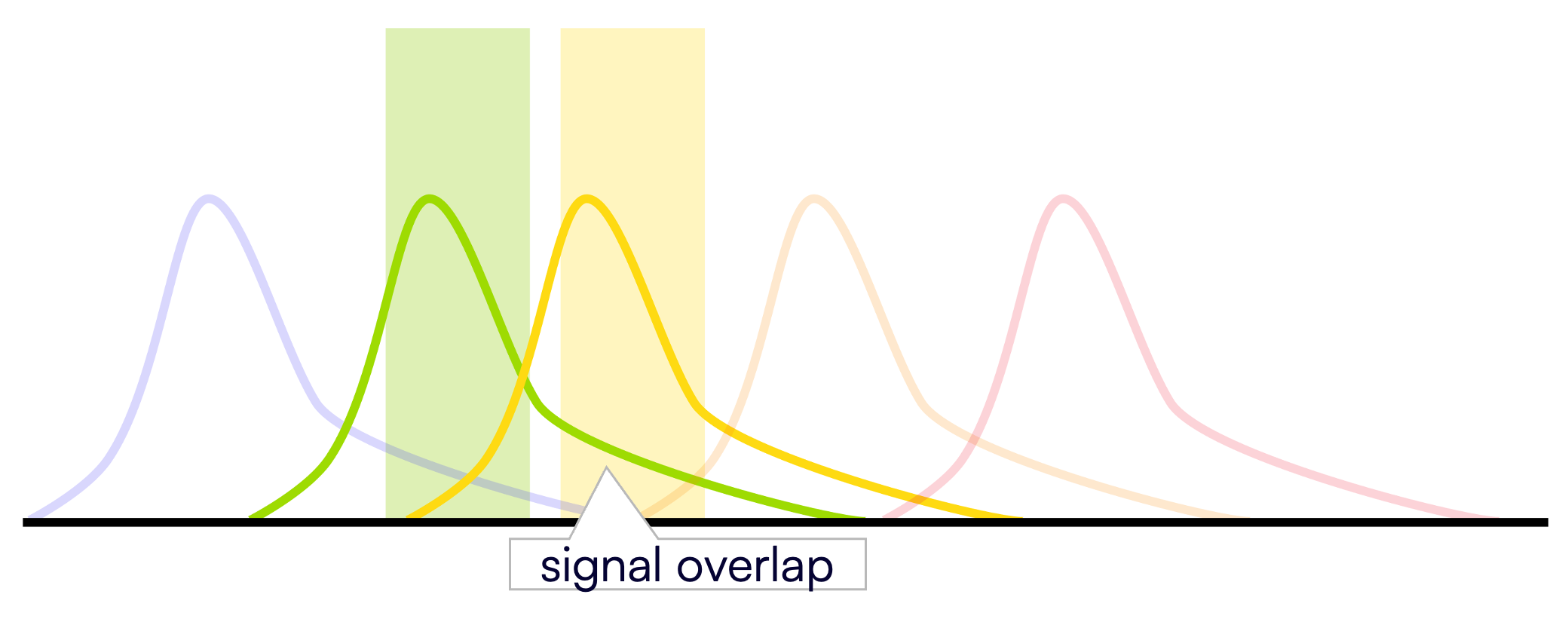
TaqMan probes require rigorous experimental design and extensive optimization to get the best signal from all targets every time you develop a new multiplex assay. Imbalance messes with your signal and makes your results difficult or even impossible to interpret.
In traditional multiplex assay design, every new target comes with the burden of probe optimization:
If you need to move quickly through discovery and analytical pipelines — particularly in cell and gene therapy — this isn't just inconvenient; it’s a slowdown on your progress.
Universal Multiplexing (UM) is as easy as adding an adapter sequence to a simple oligo pair and using pre-optimized universal probes.
Imagine multiplexing with:
The results? No more worries about tedious optimization cycles or lots of pre-experiment bioinformatics research.
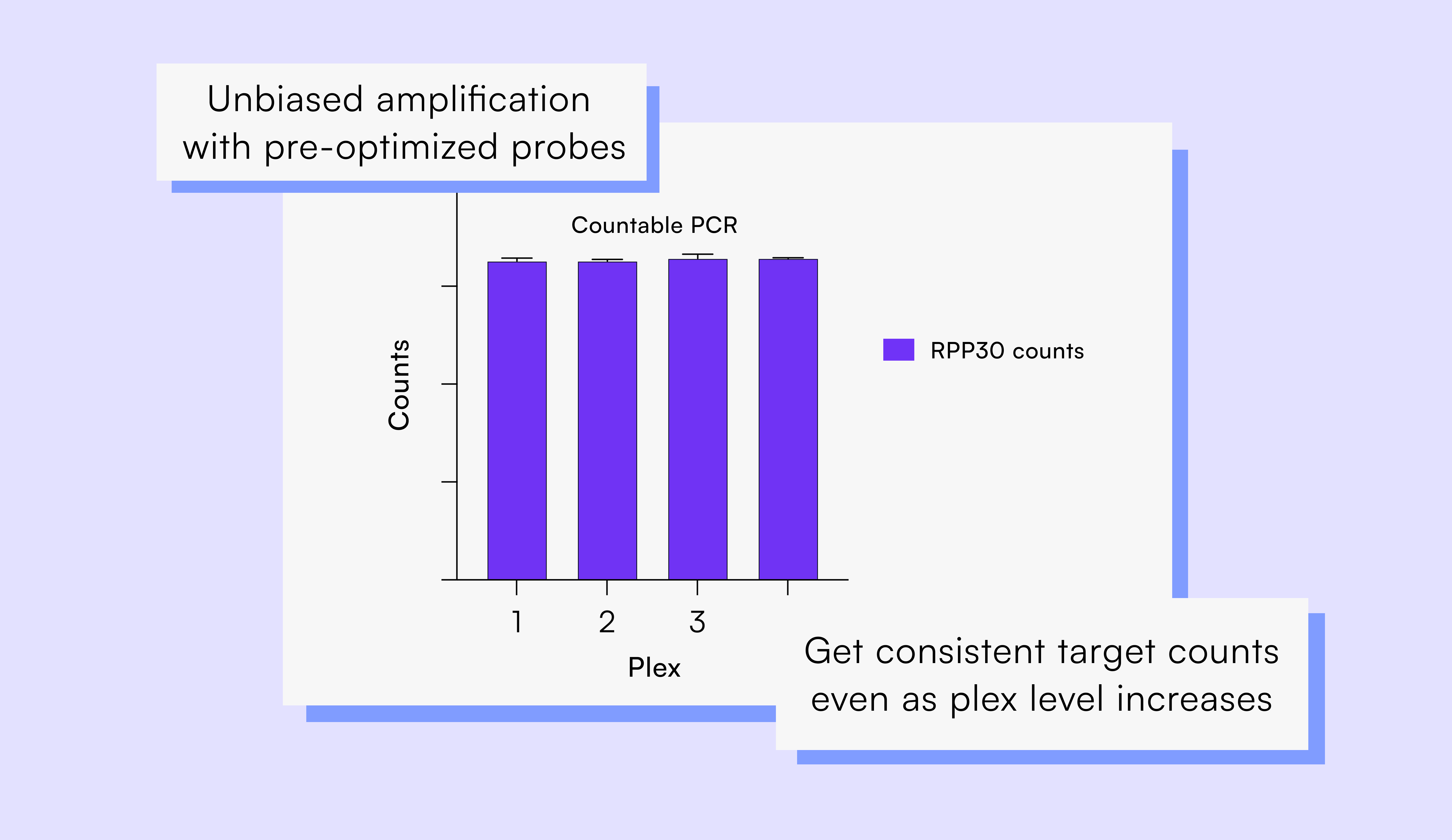
UM works out of the box. Stop the endless cycles of optimization — you’ll get a clean, crisp signal from every target, right away. No worries about signal balancing or amplification bias.

Say goodbye to amplification bias and imbalanced probe signals common to TaqMan probes on qPCR and dPCR, and say hello to exact counts that aren’t impacted by plex level.
Turn to UM and Countable PCR to end the frustration of probe optimization and get more reliable data out of every reaction.