Introduction
Precise DNA quantification is crucial for applications such as cell and gene therapy manufacturing, copy number determination, rare event detection, and general quantification of gene targets. Although digital PCR is widely used for DNA quantification, it has technical limitations that hinder low-variability results, typically yielding a coefficient of variation (CV) of around 10% [1]. These include:
- Reliance on Poisson correction, which depends on consistent and accurate partition volumes to calculate copy number reliably [2, 3, 4].
- A limited number of partitions, which begin to saturate at target counts typically near 100,000 molecules. As the target molecule counts approach the upper limit of the dynamic range, an increasing number of partitions contain multiple molecules, leading to reduced measurement resolution and accuracy [5].
- Manual thresholding, which introduces user-dependent bias and reduces reproducibility.
Countable PCR partitions individual target molecules within a 3D gel-like matrix, enabling direct counting across four channels up to 1 million counts. With over 30 million partitions ensuring single occupancy, the need for Poisson correction is eliminated, and the dependence on partition consistency and high occupancy correction is removed. In addition, Countable PCR’s light sheet imaging and automated counting algorithm allow for high-throughput processing with reduced variability between replicates and minimal user-dependent error.
Here, we demonstrate the reproducibility of Countable PCR across a six-log dynamic range and different users and instruments. These results highlight the power of direct counting to deliver low-variability data and consistent performance, which is essential for cross-validation and reliable assay development at multiple sites.
Materials and Methods
Measurement variability across a six-log dynamic range
Four short (<200 bp) synthetic double-stranded DNA templates, derived from four human genes (RPP30, JAK2, RAD51, and MET), were prepared and serially diluted to prepare counts between 10 and 1,000,000 molecules. A pool of TaqMan assays for RPP30,JAK2, RAD51, and MET were prepared for Ch01, Ch02, Ch03, and Ch04, respectively (Table 1).
Table 1. Probes and primers for reproducibility validation

The pool of synthetic templates, ranging from 10 to 1,000,000 molecules, was added to a series of multiplex PCR reactions containing the pool of TaqMan assays (Table 2).
Table 2. Four-plex Countable PCR reaction setup

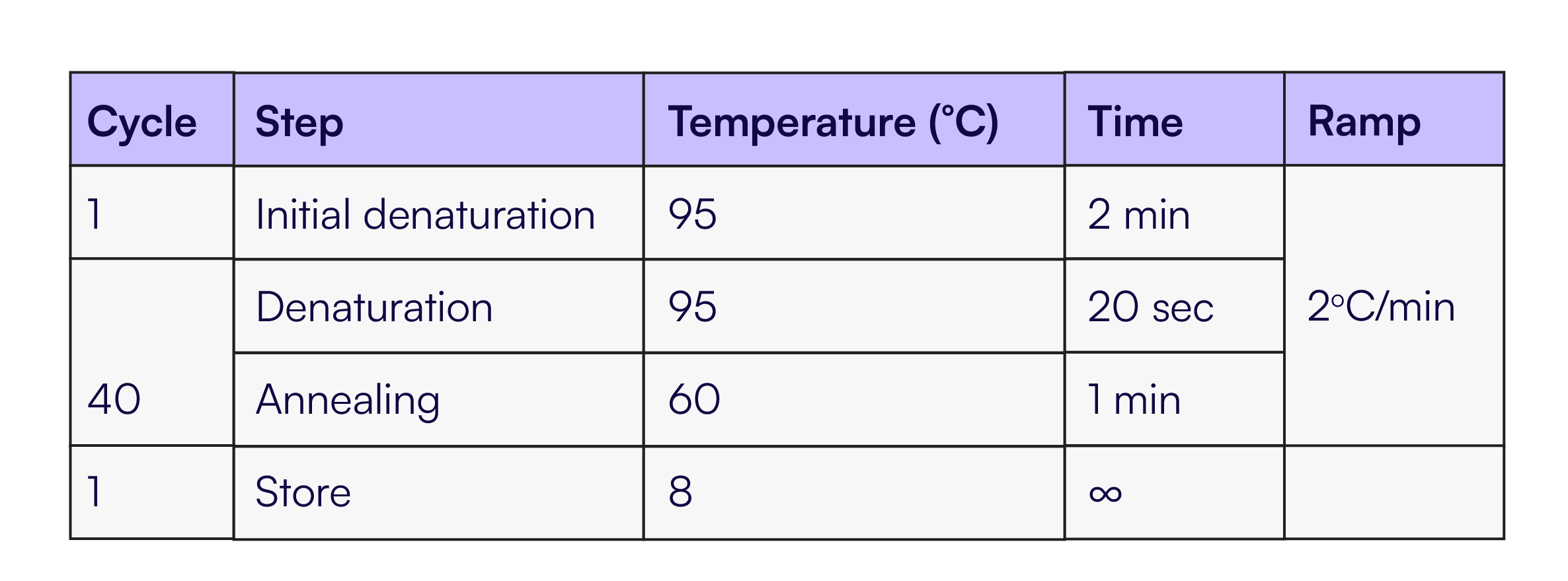
Eight replicates of each dilution were quantified with Countable PCR and used to calculate the CV at each target molecule input per channel. Thermal cycling was performed as described in Table 3.
Table 3. Thermal Cycling protocol for the RPP30, JAK2, RAD51, MET assays
Measurement variability across users and instruments
The Countable Control Assay Kit (KT0009) was used to prepare Countable PCR reactions with 100,000 counts per reaction across four channels. The kit includes primers that utilize the Universal Multiplexing chemistry to quantify four different synthetic DNA templates provided in the kit. Three users set up 12 reactions, and all 36 reactions were each quantified on three separate Countable systems. Reaction setup and thermal cycling were performed according to the Countable user guide.
Results
Countable PCR attains low CV across a six-log dynamic range in a four-plex reaction
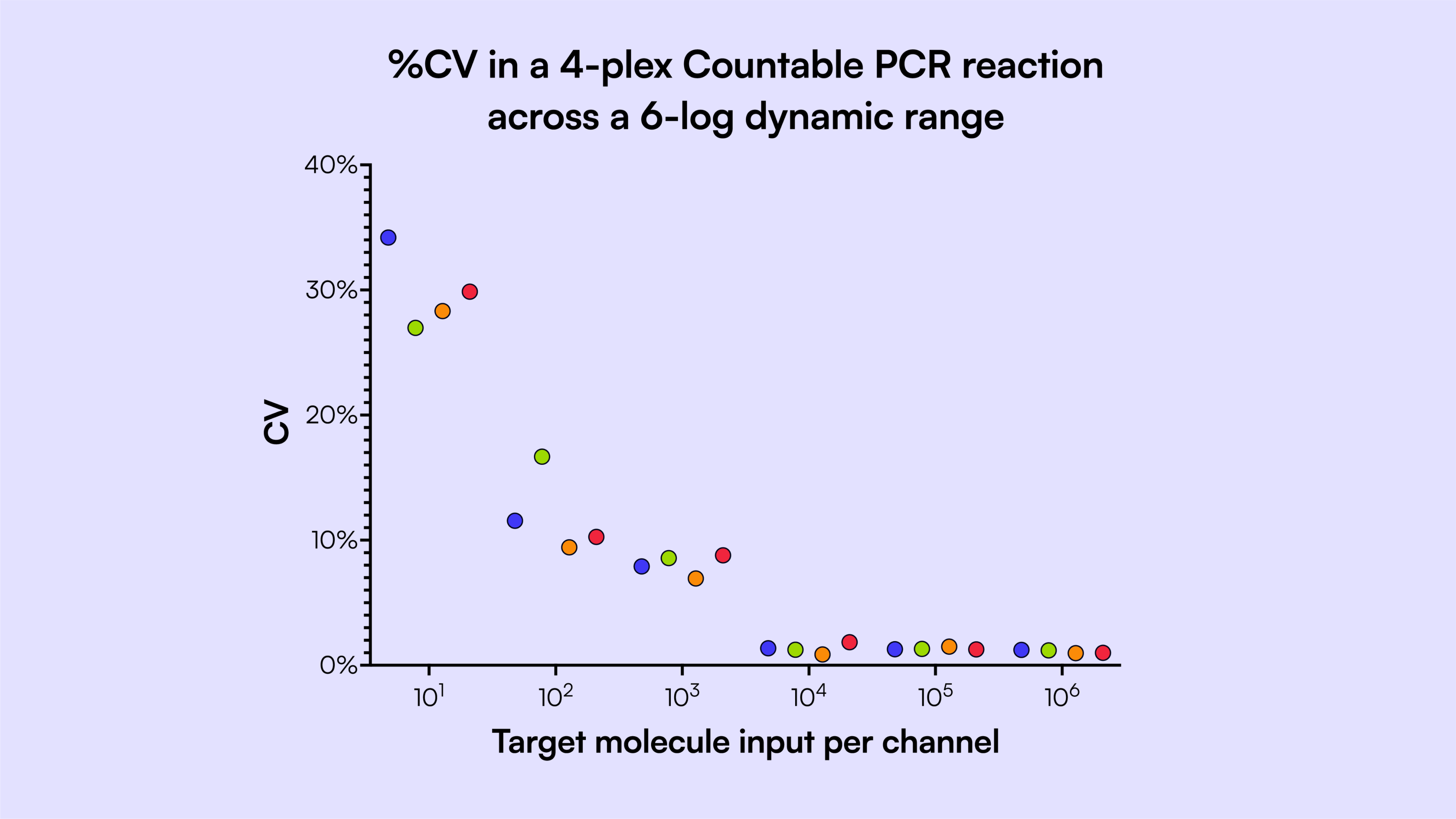
We performed Countable PCR on a series of four-plex reactions containing pooled double-stranded DNA targets RPP30, JAK2, RAD51, and MET, corresponding to Ch01, Ch02, Ch03, and Ch04, respectively. The pooled templates were serially diluted to produce final counts for each channel ranging from 10 to 1,000,000, and the CV at each concentration was calculated from eight technical replicates (Figure 1). Measurements taken at 1000 counts had a CV under 10%, and measurements taken at 10,000 counts and above had a CV under 2% across all channels.
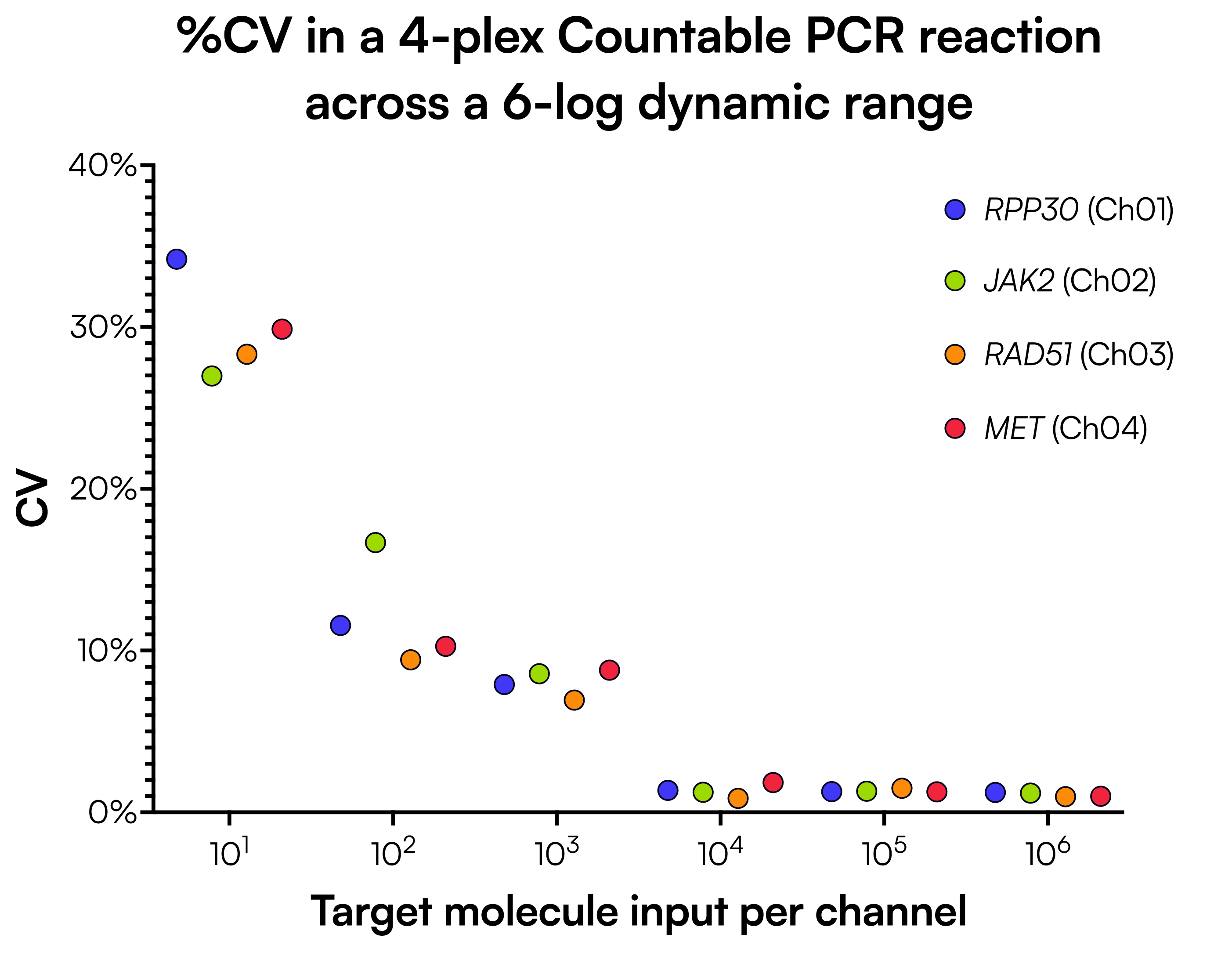
Countable PCR achieves low CV across different users and instruments
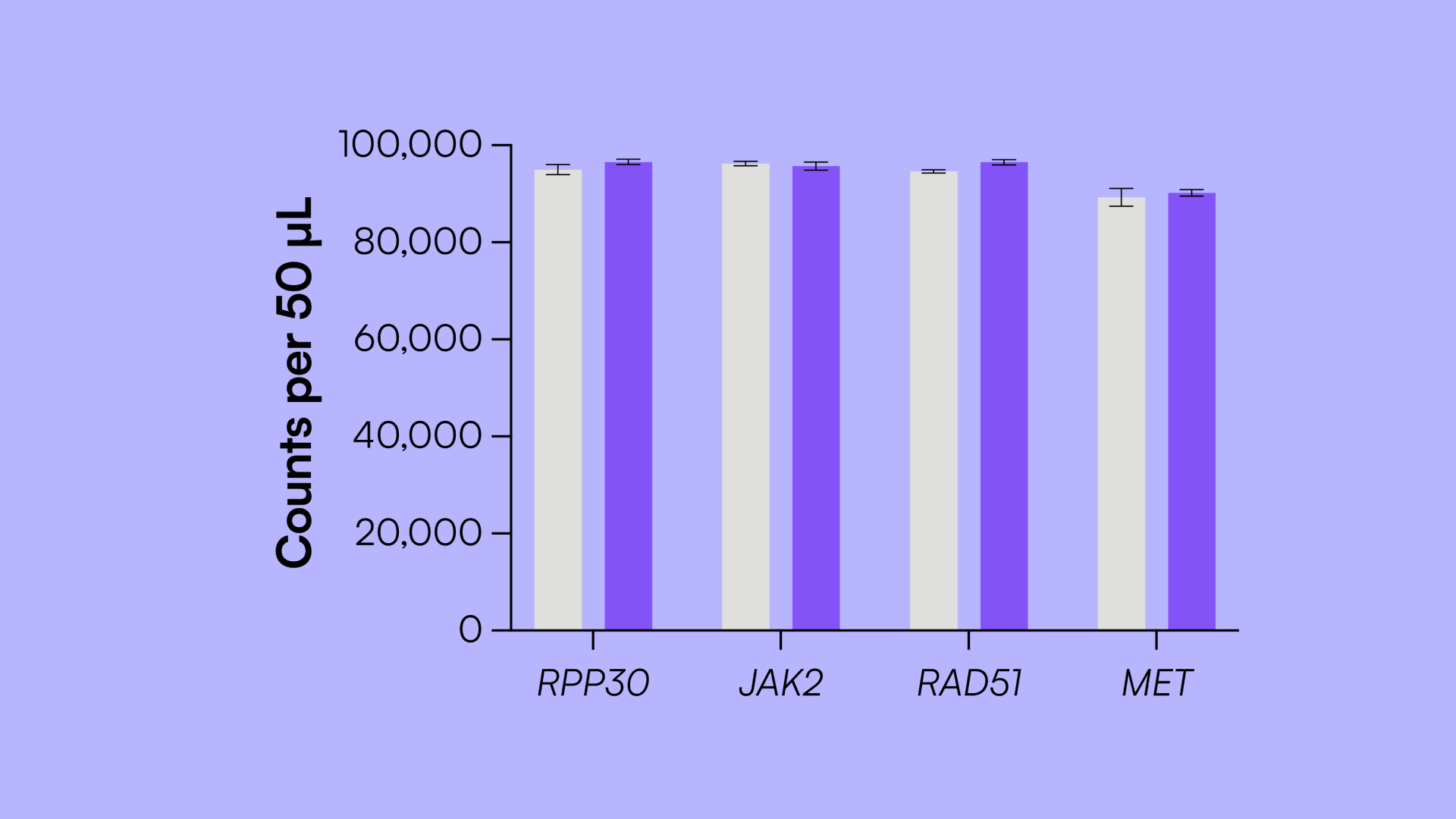
To assess measurement variability across different users and instruments, three users ran 12 reactions with the Countable Control Assay Kit and imaged each reaction on three different imagers (Table 4). Within users, the CV was very low, typically less than 1%, averaging 0.92% across all users and channels. Between users, the CV averaged 2.42% across all channels. Between three different instruments, the CV averaged 0.62% across all channels and users. These results demonstrate strong concordance of data across users and Countable PCR systems, supporting their use in cross-functional workflows and cross-site collaborations.
Table 4: The Countable system is highly reproducible between technical replicates within each user, across instruments, and between users.

Conclusion
Countable PCR delivers results with exceptionally low variability across technical replicates, users, and systems across a wide dynamic range. Specifically, this study demonstrates:
- Exceptional precision among technical replicates, with CVs below 1% for measurements around 100,000 counts.
- High reproducibility among users, with CVs below 3%.
- High reproducibility across instruments, with a CV of 0.65% across three systems.
These highly reproducible results highlight Countable PCR’s robustness as a platform for quantifying gene targets at multiple points within a workflow and for cross-operator or cross-site experiment validation.
References
1. Tumpach C. et al., Adaptation of the intact proviral DNA assay to a nanowell-based digital PCR platform. J Virus Erad. 2023 June, 9(2), 100335
2. Bhat, S. et al., Single molecule detection in nanofluidic digital array enables accurate measurement of DNA copy number. Anal Bioanal Chem. 2009 March, 394, 457-4
3. Corbisier, P. et al., DNA copy number concentration measured by digital and droplet digital quantitative PCR using certified reference materials. Anal Bioanal Chem. 2015 March, 407(7), 1831-1840
4. Kosir, AB. et al., Droplet volume variability as a critical factor for accuracy of absolute quantification using droplet digital PCR. Anal Bioanal Chem. 2017 November, 409(28), 6689-6697
5. Majumdar, N. et al., Digital PCR Modeling for Maximal Sensitivity, Dynamic Range and Measurement Precision. PloS one. 2015 March, 10(3), e0118833
Introduction
Precise DNA quantification is crucial for applications such as cell and gene therapy manufacturing, copy number determination, rare event detection, and general quantification of gene targets. Although digital PCR is widely used for DNA quantification, it has technical limitations that hinder low-variability results, typically yielding a coefficient of variation (CV) of around 10% [1]. These include:
- Reliance on Poisson correction, which depends on consistent and accurate partition volumes to calculate copy number reliably [2, 3, 4].
- A limited number of partitions, which begin to saturate at target counts typically near 100,000 molecules. As the target molecule counts approach the upper limit of the dynamic range, an increasing number of partitions contain multiple molecules, leading to reduced measurement resolution and accuracy [5].
- Manual thresholding, which introduces user-dependent bias and reduces reproducibility.
Countable PCR partitions individual target molecules within a 3D gel-like matrix, enabling direct counting across four channels up to 1 million counts. With over 30 million partitions ensuring single occupancy, the need for Poisson correction is eliminated, and the dependence on partition consistency and high occupancy correction is removed. In addition, Countable PCR’s light sheet imaging and automated counting algorithm allow for high-throughput processing with reduced variability between replicates and minimal user-dependent error.
Here, we demonstrate the reproducibility of Countable PCR across a six-log dynamic range and different users and instruments. These results highlight the power of direct counting to deliver low-variability data and consistent performance, which is essential for cross-validation and reliable assay development at multiple sites.
Materials and Methods
Measurement variability across a six-log dynamic range
Four short (<200 bp) synthetic double-stranded DNA templates, derived from four human genes (RPP30, JAK2, RAD51, and MET), were prepared and serially diluted to prepare counts between 10 and 1,000,000 molecules. A pool of TaqMan assays for RPP30,JAK2, RAD51, and MET were prepared for Ch01, Ch02, Ch03, and Ch04, respectively (Table 1).
Table 1. Probes and primers for reproducibility validation

The pool of synthetic templates, ranging from 10 to 1,000,000 molecules, was added to a series of multiplex PCR reactions containing the pool of TaqMan assays (Table 2).
Table 2. Four-plex Countable PCR reaction setup

Eight replicates of each dilution were quantified with Countable PCR and used to calculate the CV at each target molecule input per channel. Thermal cycling was performed as described in Table 3.
Table 3. Thermal Cycling protocol for the RPP30, JAK2, RAD51, MET assays

Measurement variability across users and instruments
The Countable Control Assay Kit (KT0009) was used to prepare Countable PCR reactions with 100,000 counts per reaction across four channels. The kit includes primers that utilize the Universal Multiplexing chemistry to quantify four different synthetic DNA templates provided in the kit. Three users set up 12 reactions, and all 36 reactions were each quantified on three separate Countable systems. Reaction setup and thermal cycling were performed according to the Countable user guide.
Results
Countable PCR attains low CV across a six-log dynamic range in a four-plex reaction
We performed Countable PCR on a series of four-plex reactions containing pooled double-stranded DNA targets RPP30, JAK2, RAD51, and MET, corresponding to Ch01, Ch02, Ch03, and Ch04, respectively. The pooled templates were serially diluted to produce final counts for each channel ranging from 10 to 1,000,000, and the CV at each concentration was calculated from eight technical replicates (Figure 1). Measurements taken at 1000 counts had a CV under 10%, and measurements taken at 10,000 counts and above had a CV under 2% across all channels.
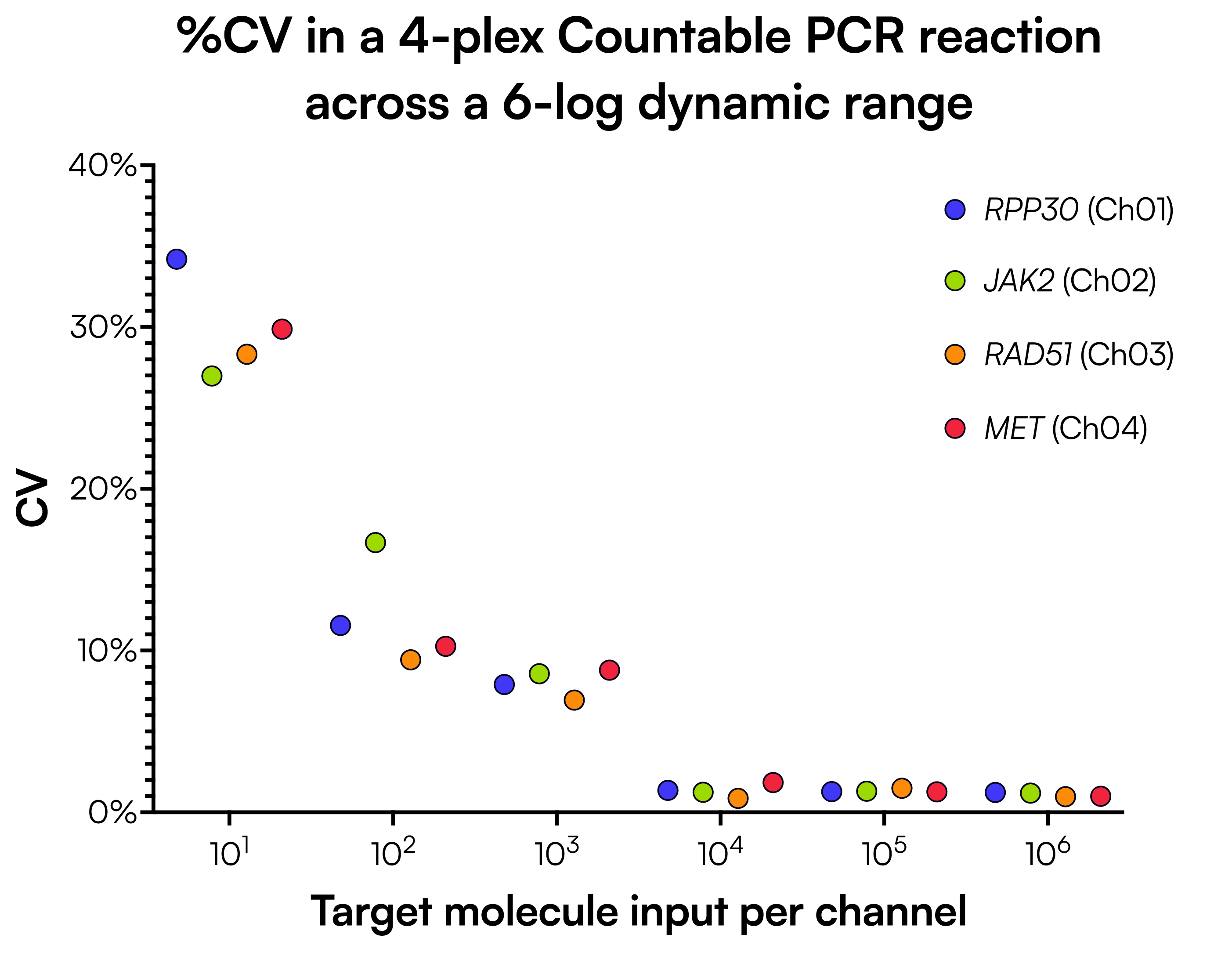
Countable PCR achieves low CV across different users and instruments
To assess measurement variability across different users and instruments, three users ran 12 reactions with the Countable Control Assay Kit and imaged each reaction on three different imagers (Table 4). Within users, the CV was very low, typically less than 1%, averaging 0.92% across all users and channels. Between users, the CV averaged 2.42% across all channels. Between three different instruments, the CV averaged 0.62% across all channels and users. These results demonstrate strong concordance of data across users and Countable PCR systems, supporting their use in cross-functional workflows and cross-site collaborations.
Table 4: The Countable system is highly reproducible between technical replicates within each user, across instruments, and between users.

Conclusion
Countable PCR delivers results with exceptionally low variability across technical replicates, users, and systems across a wide dynamic range. Specifically, this study demonstrates:
- Exceptional precision among technical replicates, with CVs below 1% for measurements around 100,000 counts.
- High reproducibility among users, with CVs below 3%.
- High reproducibility across instruments, with a CV of 0.65% across three systems.
These highly reproducible results highlight Countable PCR’s robustness as a platform for quantifying gene targets at multiple points within a workflow and for cross-operator or cross-site experiment validation.
References
1. Tumpach C. et al., Adaptation of the intact proviral DNA assay to a nanowell-based digital PCR platform. J Virus Erad. 2023 June, 9(2), 100335
2. Bhat, S. et al., Single molecule detection in nanofluidic digital array enables accurate measurement of DNA copy number. Anal Bioanal Chem. 2009 March, 394, 457-4
3. Corbisier, P. et al., DNA copy number concentration measured by digital and droplet digital quantitative PCR using certified reference materials. Anal Bioanal Chem. 2015 March, 407(7), 1831-1840
4. Kosir, AB. et al., Droplet volume variability as a critical factor for accuracy of absolute quantification using droplet digital PCR. Anal Bioanal Chem. 2017 November, 409(28), 6689-6697
5. Majumdar, N. et al., Digital PCR Modeling for Maximal Sensitivity, Dynamic Range and Measurement Precision. PloS one. 2015 March, 10(3), e0118833


.png)