Introduction
Multiplex PCR is a powerful technique that enables the simultaneous detection of multiple targets in a single reaction [1]. To date, the development of multiplex PCR assays has often presented challenges that have limited their widespread adoption. These challenges revolve largely around:
- Optimization of primer/probe pairs to avoid amplification bias and artifacts
- Prohibitive cost of scaling experiments with custom hydrolysis probes (HP) such as TaqMan (TM) probes
Countable PCR isolates each DNA molecule in its compartment within a gel-like matrix for independent amplification [2]. Multiplexing is achievable with minimal optimization as there is no competition between targets within compartments. This alleviates the need for many rounds of optimization.
To overcome challenges related to cost, Countable Labs developed Universal Multiplexing (UM), a novel chemistry that eliminates the need for target-specific HPs. UM uses generic prefixed probe sequences with target-specific primers. Compared to HP-based approaches, UM significantly reduces assay design and run costs by eliminating expensive target-specific probes that require lengthy synthesis times [3, 4, 5].
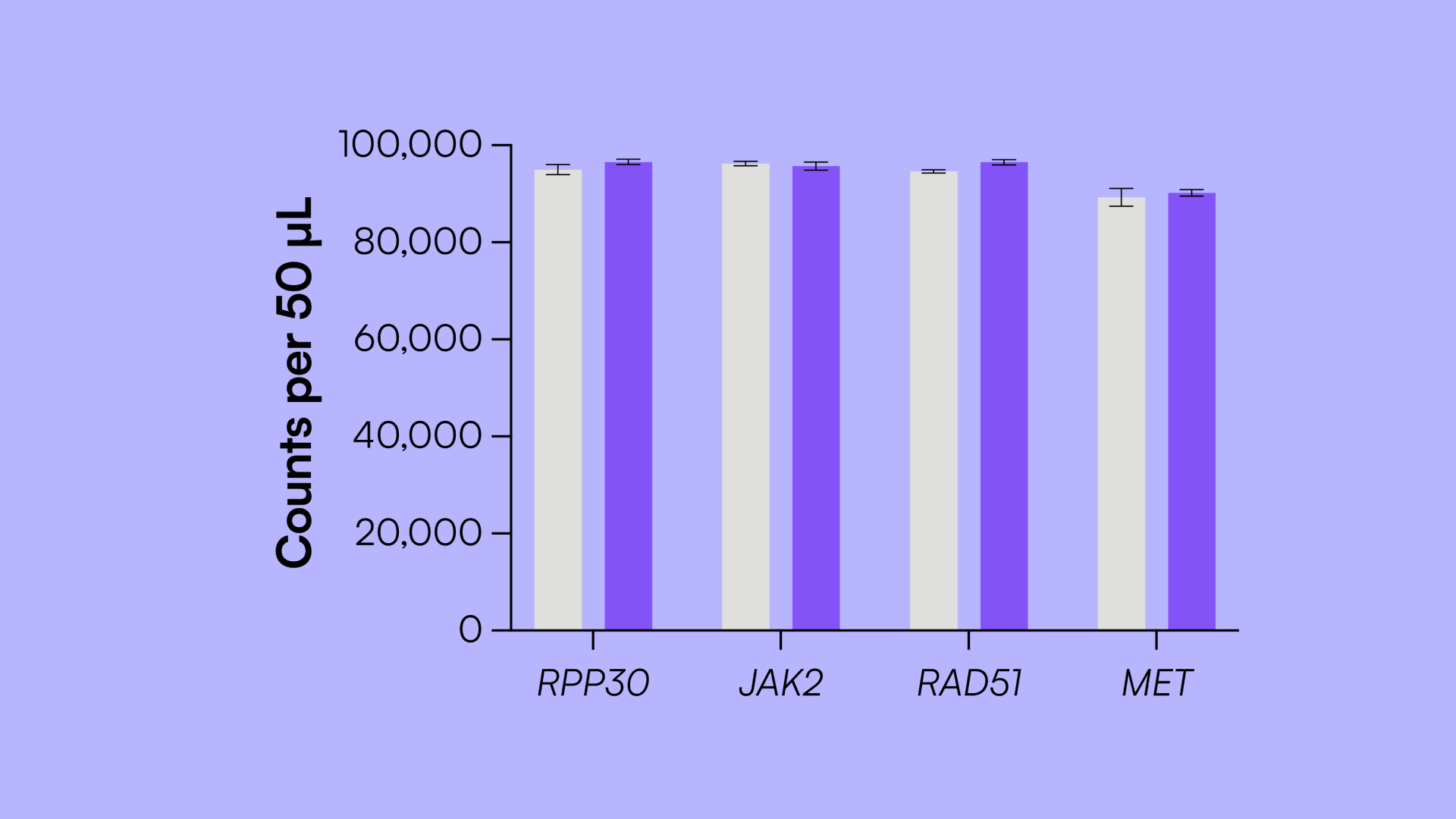
In this study, we demonstrate the development of a 4-plex UM assay based on an assay originally developed using HP, and compare the performance of both on the Countable PCR platform.
Introduction
Multiplex PCR is a powerful technique that enables the simultaneous detection of multiple targets in a single reaction [1]. To date, the development of multiplex PCR assays has often presented challenges that have limited their widespread adoption. These challenges revolve largely around:
- Optimization of primer/probe pairs to avoid amplification bias and artifacts
- Prohibitive cost of scaling experiments with custom hydrolysis probes (HP) such as TaqMan (TM) probes
Countable PCR isolates each DNA molecule in its compartment within a gel-like matrix for independent amplification [2]. Multiplexing is achievable with minimal optimization as there is no competition between targets within compartments. This alleviates the need for many rounds of optimization.
To overcome challenges related to cost, Countable Labs developed Universal Multiplexing (UM), a novel chemistry that eliminates the need for target-specific HPs. UM uses generic prefixed probe sequences with target-specific primers. Compared to HP-based approaches, UM significantly reduces assay design and run costs by eliminating expensive target-specific probes that require lengthy synthesis times [3, 4, 5].
In this study, we demonstrate the development of a 4-plex UM assay based on an assay originally developed using HP, and compare the performance of both on the Countable PCR platform.