
Abstract
Quantitative or digital PCR is commonly used to validate biomarkers following RNA-seq studies, often involving multiple gene markers. PCR measurement of panels of gene expression markers also forms the basis for various clinical diagnostic and prognostic tests.Multiplexing, combining several biomarkers in a single PCR reaction, improves efficiency and accuracy while reducing time, reagent use, and sample consumption. However, designing high-order multiplex PCR panels typically requires extensive optimization and iterative assay development. Having faster,more straightforward approaches to develop multiplexed PCR panels is highly desirable.
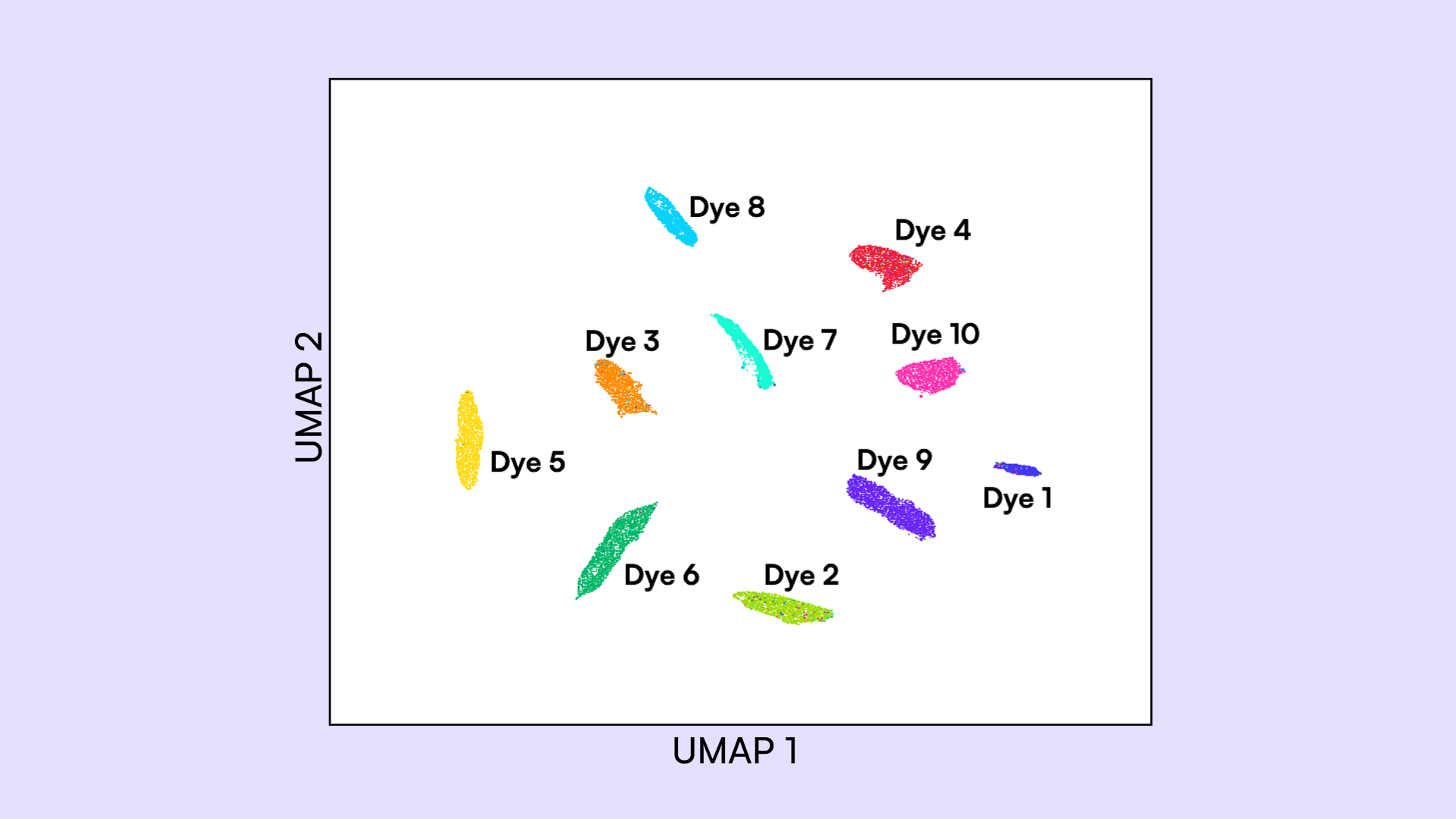
A major technical challenge of high-order multiplexing is balancing amplification efficiency across targets and resolving spectral overlaps of fluorophores used to detect each target. To overcome these challenges and open up new, more straightforward avenues for high-order multiplexing, we utilized Countable PCR, a single-molecule DNA quantification technique. Using the 4-color Countable system, we successfully resolved 10 distinct fluorescent dyes in one PCR reaction. By applying combinatorial labeling with 8 fluorophores, we developed a 16-target gene expression panel with marker genes specific to various human tissues.
This high-precision, high-order multiplexing method significantly expands the capabilities of PCR, dramatically reducing panel development time and accelerating validation studies following bulk- or single-cell RNA-seq. Further, it has the potential to eliminate or reduce the need for NGS in many biomarker quantification applications. Beyond validation studies, this approach holds strong potential for clinical assays that are based on panels of gene expression markers.
Abstract
Quantitative or digital PCR is commonly used to validate biomarkers following RNA-seq studies, often involving multiple gene markers. PCR measurement of panels of gene expression markers also forms the basis for various clinical diagnostic and prognostic tests.Multiplexing, combining several biomarkers in a single PCR reaction, improves efficiency and accuracy while reducing time, reagent use, and sample consumption. However, designing high-order multiplex PCR panels typically requires extensive optimization and iterative assay development. Having faster,more straightforward approaches to develop multiplexed PCR panels is highly desirable.
A major technical challenge of high-order multiplexing is balancing amplification efficiency across targets and resolving spectral overlaps of fluorophores used to detect each target. To overcome these challenges and open up new, more straightforward avenues for high-order multiplexing, we utilized Countable PCR, a single-molecule DNA quantification technique. Using the 4-color Countable system, we successfully resolved 10 distinct fluorescent dyes in one PCR reaction. By applying combinatorial labeling with 8 fluorophores, we developed a 16-target gene expression panel with marker genes specific to various human tissues.
This high-precision, high-order multiplexing method significantly expands the capabilities of PCR, dramatically reducing panel development time and accelerating validation studies following bulk- or single-cell RNA-seq. Further, it has the potential to eliminate or reduce the need for NGS in many biomarker quantification applications. Beyond validation studies, this approach holds strong potential for clinical assays that are based on panels of gene expression markers.

