
Abstract
BRAF is a critical gene of interest due to its implication in many cancers,including thyroid, melanoma, and colorectal cancer. Specifically,clinically-relevant mutations V600E, V600K, and V600R have been identified as the most common oncogenic mutations, and the presence of single- or double-point mutations in patients are important factors in therapeutic decision-making.
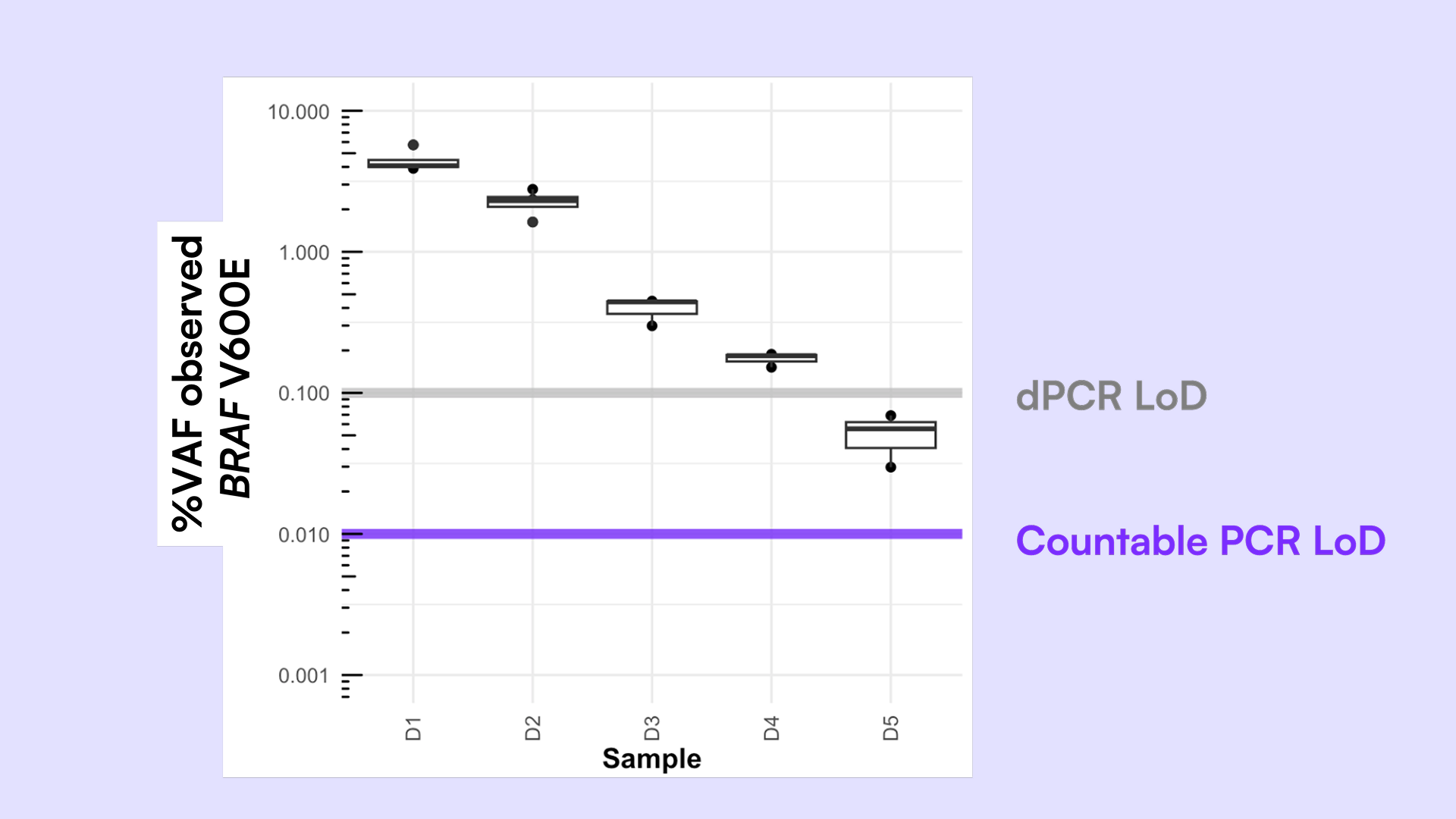
Accurate detection of BRAF mutations in liquid biopsy samples remains challenging due to the limited sample available for analysis and the difficulty of distinguishing closely related variants. Conventional qPCR and digital PCR (dPCR) approaches are constrained by low input tolerance, limited dynamic range, and poor scalability for multiplexed mutation detection. Due to these technical limitations, these approaches are limited to VAF% (variable allele frequency)of about 1-5% for qPCR and 0.1% for dPCR, but researchers demand more sensitive and faster VAF% determination as therapeutic treatments evolve.
Here,we describe a one-tube, 4-plex PCR assay capable of distinguishing and quantifying BRAF mutations with high sensitivity via single-molecule counting.With the method introduced here, combined with an optional pre-amplification step, the assay achieves a 6-log dynamic range, enabling direct comparison of WT and low abundance mutants. Furthermore, with no dead volume, it was possible to directly amplify low-abundance targets from 35 μL of input sample, effectively increasing the sensitivity of the methodology.
These abilities allowed us to improve limit of detection (LoD) and confidence in the results, demonstrating detection of rare variants down to <0.05% VAF. The ability to measure all common BRAF mutations in one tube preserves precious samples.
Abstract
BRAF is a critical gene of interest due to its implication in many cancers,including thyroid, melanoma, and colorectal cancer. Specifically,clinically-relevant mutations V600E, V600K, and V600R have been identified as the most common oncogenic mutations, and the presence of single- or double-point mutations in patients are important factors in therapeutic decision-making.
Accurate detection of BRAF mutations in liquid biopsy samples remains challenging due to the limited sample available for analysis and the difficulty of distinguishing closely related variants. Conventional qPCR and digital PCR (dPCR) approaches are constrained by low input tolerance, limited dynamic range, and poor scalability for multiplexed mutation detection. Due to these technical limitations, these approaches are limited to VAF% (variable allele frequency)of about 1-5% for qPCR and 0.1% for dPCR, but researchers demand more sensitive and faster VAF% determination as therapeutic treatments evolve.
Here,we describe a one-tube, 4-plex PCR assay capable of distinguishing and quantifying BRAF mutations with high sensitivity via single-molecule counting.With the method introduced here, combined with an optional pre-amplification step, the assay achieves a 6-log dynamic range, enabling direct comparison of WT and low abundance mutants. Furthermore, with no dead volume, it was possible to directly amplify low-abundance targets from 35 μL of input sample, effectively increasing the sensitivity of the methodology.
These abilities allowed us to improve limit of detection (LoD) and confidence in the results, demonstrating detection of rare variants down to <0.05% VAF. The ability to measure all common BRAF mutations in one tube preserves precious samples.

