Abstract
HPV-positive oropharyngeal squamous cell carcinomas are rising, yet detection often occurs in later stages of disease due to lack of early-stage symptoms. Currently, P16 immunohistochemistry is the standard for detection, but it is not a direct measurement of HPV presence; further, assessment of P16 levels is subjective, lacks strain specificity, and yields equivocal results in approximately 10% of cases. These often require slow follow-up testing such asin situ hybridization. A faster, subtype-resolved method with better objectivity would be an improvement over current technologies.
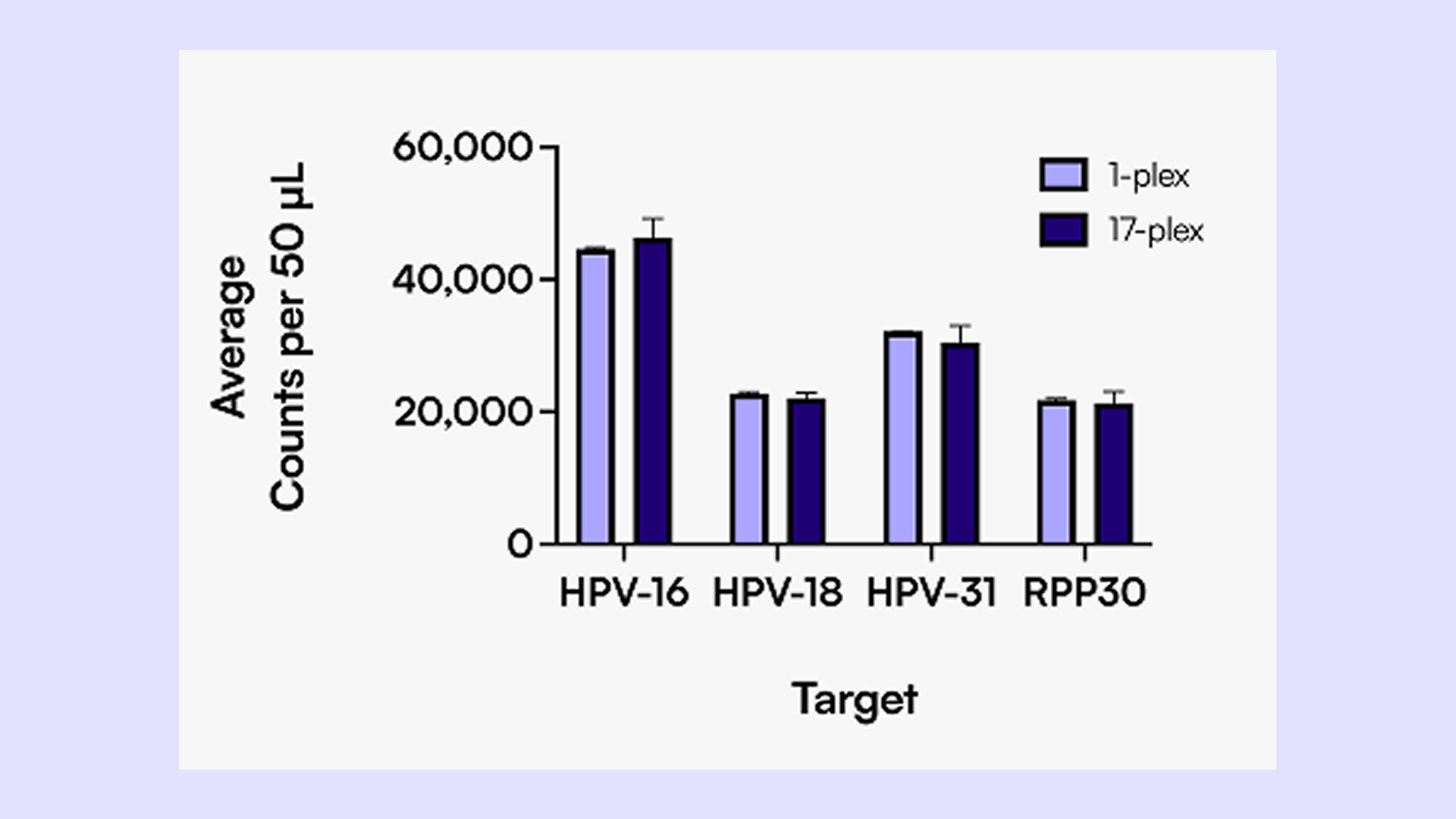
We utilized Countable PCR, a PCR technique that enables direct counting of single molecules across a dynamic range up to 106, to develop a 17-plex panel that differentiates and quantifies: the 2 highest-risk HPV strains for oropharyngeal cancer (HPV-16, HPV-18), 1 common region for 14 high-risk subtypes in parallel (HPV- 16, 18, 31, 33, 35, 39, 45, 51, 51, 56, 58, 59, 66, and 68), and 1 internal control (RPP30) with high precision and sensitivity. Samples categorized as either positive, negative, or equivocal based on p16 immunostaining at USC were re-tested using Countable PCR.
The broad dynamic range of Countable PCR enabled minimal optimization for the panel, even in samples containing high HPV load. Further, Countable PCR was able to resolve equivocal samples in just a few hours, rather than the days required forin situ hybridization. Use of this Countable PCR 17-plex panel also allowed identification of all HPV subtypes while existing protocols based on real-time PCR can only identify HPV-16 and HPV-18 while lumping all other high-risk subtypes as a single entity.
This proof-of-concept validation experiment demonstrates the value of a quantitative detection panel thatis able todelineate HPV infection across multiple subtypes within a few hours.
Abstract
HPV-positive oropharyngeal squamous cell carcinomas are rising, yet detection often occurs in later stages of disease due to lack of early-stage symptoms. Currently, P16 immunohistochemistry is the standard for detection, but it is not a direct measurement of HPV presence; further, assessment of P16 levels is subjective, lacks strain specificity, and yields equivocal results in approximately 10% of cases. These often require slow follow-up testing such asin situ hybridization. A faster, subtype-resolved method with better objectivity would be an improvement over current technologies.
We utilized Countable PCR, a PCR technique that enables direct counting of single molecules across a dynamic range up to 106, to develop a 17-plex panel that differentiates and quantifies: the 2 highest-risk HPV strains for oropharyngeal cancer (HPV-16, HPV-18), 1 common region for 14 high-risk subtypes in parallel (HPV- 16, 18, 31, 33, 35, 39, 45, 51, 51, 56, 58, 59, 66, and 68), and 1 internal control (RPP30) with high precision and sensitivity. Samples categorized as either positive, negative, or equivocal based on p16 immunostaining at USC were re-tested using Countable PCR.
The broad dynamic range of Countable PCR enabled minimal optimization for the panel, even in samples containing high HPV load. Further, Countable PCR was able to resolve equivocal samples in just a few hours, rather than the days required forin situ hybridization. Use of this Countable PCR 17-plex panel also allowed identification of all HPV subtypes while existing protocols based on real-time PCR can only identify HPV-16 and HPV-18 while lumping all other high-risk subtypes as a single entity.
This proof-of-concept validation experiment demonstrates the value of a quantitative detection panel thatis able todelineate HPV infection across multiple subtypes within a few hours.