Rain belongs to the weather, not your PCR data – how Countable PCR delivers clear data


Human biology is complex — with over 20,000 known genes for regulation and protein expression, there’s a lot to investigate. So, you need to design multiplex experiments — a more accurate reflection of the real biology.
dPCR promised to make this achievable, but it still takes weeks or even months to optimize multiplex assays. And the culprit is rain.
Rain leads to costly and time-consuming experimental redesign and optimization — and is the reason your multiplex assays are slowing down your lab work.
But Countable PCR doesn’t have the same technical limitations as dPCR, so it’s clear skies ahead.
Rain is the result of uneven amplification within dPCR partitions. It occurs for a variety of reasons:

Figure 1: Inefficient amplification for a single target. Here, the effects of inefficient amplification and insufficient partitioning add up (left panel) to skew the count estimated by Poisson after digital signal conversion (right panel)

Figure 2: Amplification bias impacts multiplexing in partitions. In this example, multiple targets occupy a single partition, and the blue target has better amplification efficiency than orange (left panel). The result is systematic undercounting of the orange target after digital signal conversion (right panel).
When multiplexing, there should only be a single target in each partition to reduce this competition – but that’s not always possible, and you don’t know for sure.
Now, it’s up to you to set a threshold to decide which populations are “1” and which are “0” so Poisson distribution will apply. And your data looks like Figure 3.
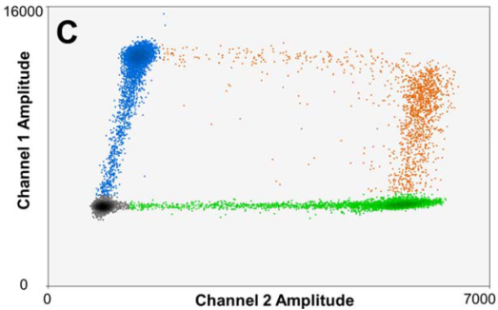
Figure 3: Sample data from a 2-plex digital PCR experiment demonstrating extensive rain for single- and double-plexed signals. For each partition: black signal = no targets; blue = target 1 only; green = target 2 only; orange = target 1 + target 2.
See the long tails between the populations – where’s the cutoff between each? It’s not easy to spot.
And the more targets you have, the more this amplification bias becomes an issue.
Rainy dPCR data isn’t just casting doubt on your results, it’s also a barrier to regulatory agency approval.
So, you’re forced to do lots of optimization to reduce the rainy signal.
Each target + probe combo in multiplex is unique, and it takes weeks or even months to solve the rain problem.
And for each target you look at, the problem scales. Lots of scientists don’t bother with more than 2-plex experiments in dPCR because it’s not worth the stress.
Countable PCR means no concerns about rain – ever.
With true single-molecule occupancy, there aren’t concerns about amplification bias:

Figure 4: 4-plex signal from a representative light sheet in Countable PCR. Each target occupies a single compartment, so they do not compete for amplification reagents and give unbiased amplification.
You’ll get clean, crisp signal from up to 4 channels that eliminates months of optimization. So, you’re able to probe deeper into the real biology of your sample with a lot less stress.
Experience the difference Countable PCR makes for multi-gene target analysis.