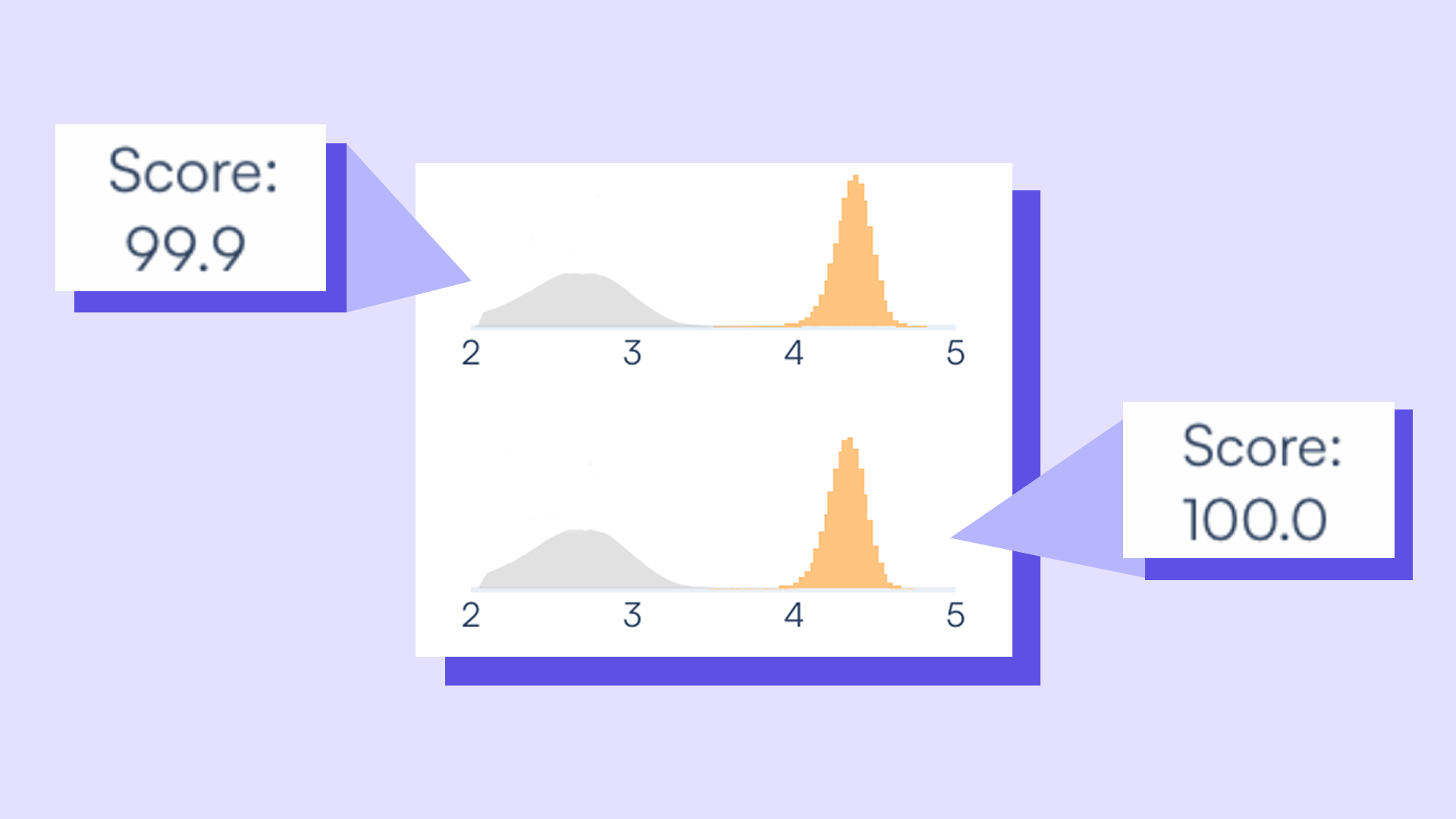

In rare event detection, time is tight – and often, your sample is limited. So why are you splitting it five times to get one answer?
Hyperwelling is the tedious process of splitting one sample across multiple runs. It’s become standard practice in digital PCR because of technical limitations. But it’s not efficient, it’s not scalable, and it’s not necessary anymore, thanks to Countable PCR.
Hyperwelling isn’t just annoying, it’s expensive
Every extra run requires more consumables, more labor, and more time from assay setup to insights. In high-throughput settings like CLIA labs, that adds up fast.
Countable PCR gets to the answers with less work and less material. A single 50 µL reaction gets you the data you need — faster, simpler, and with more confidence in your result.
The dPCR problem: too little volume, too many workarounds
Here’s what’s happening behind the scenes with dPCR:
- Most systems only allow for 5–25 µL of input volume — and in systems that rely on microfluidics, as much as half of that is lost to fluidics processing
- Once your target counts exceed about 10⁵, Poisson statistics fall apart, giving you a noisy readout and questionable data
The workaround? Split your sample – and therefore your rare events – and run it across multiple wells to gain statistical confidence. This means one sample has now become three or even five experiments, which represent just one technical replicate.
When you consider the need to include technical replicates, you realize how dPCR becomes costly fast when you’re trying to hunt for rare molecules.
.png)
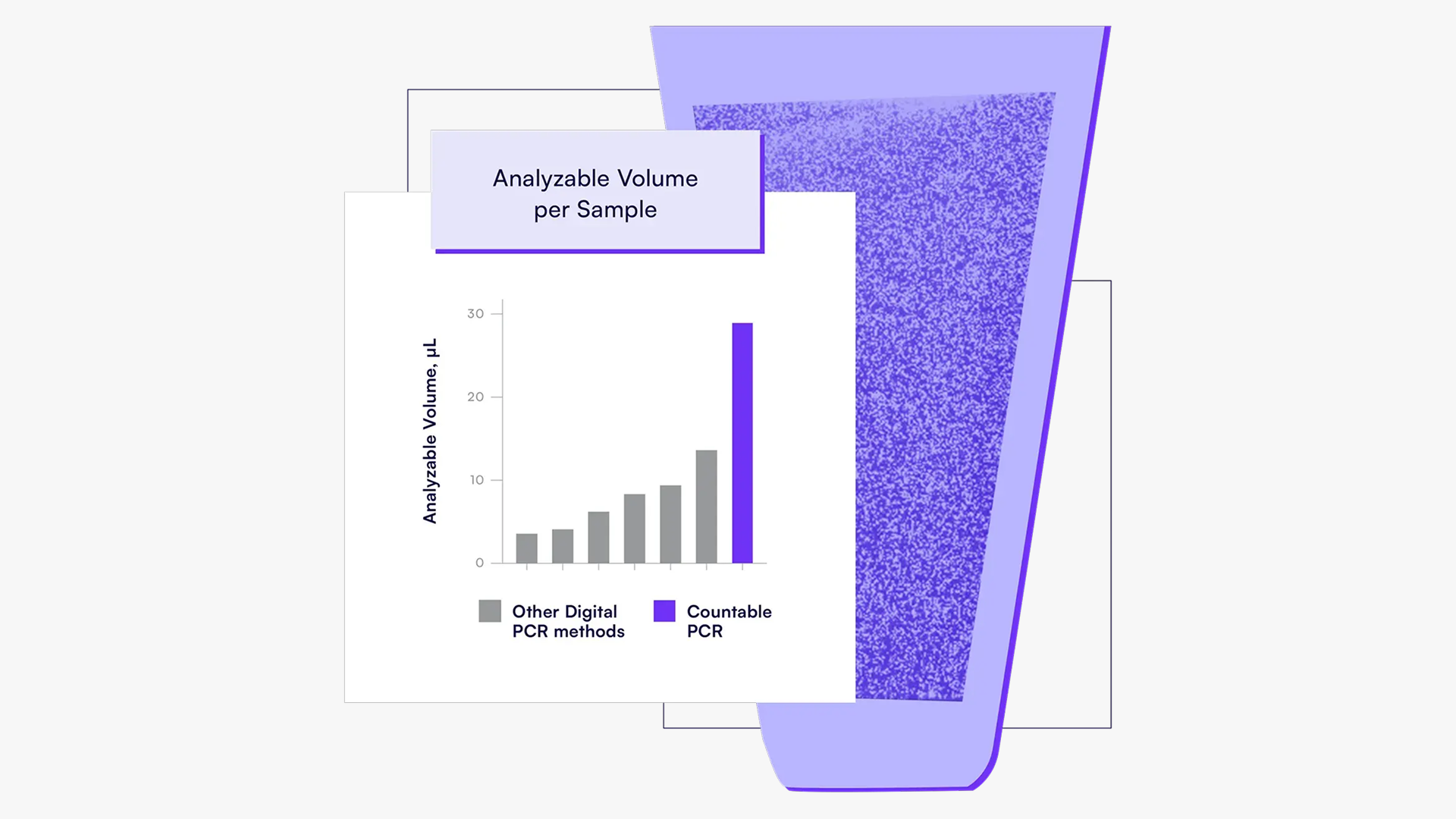
Do you know how much of your sample volume is actually analyzed by your PCR system? Here's a comparison between Countable PCR and various other PCR systems, based on the volume that gets analyzed. With Countable PCR, 100% of the input volume is analyzed. In contrast, microfluidics-based digital PCR systems may analyze as little as 50% of what’s added to the wells.
Load everything into one reaction, feel confident in your data
Countable PCR ditches the need for statistical gymnastics.
Instead of estimating based on partitions and probabilities, you count directly — across more than 30 million single-molecule compartments. That gives you 6-log dynamic range, which lets you work with the whole picture, not pieces of it.
- Load up to 35 µL or 1 µg of DNA in a single tube — perfect when doing rare event detection on dilute samples like liquid biopsy. No more wasting precious patient samples
- Count across a 6-log dynamic range — so you capture rare and common targets side by side, pick up more of your rare events for statistical confidence, while still being able to measure the WT or benchmarking gene you’re comparing against
- Get to 0.004% VAF in one reaction — with no hyperwelling or experimental workarounds
No wasted volume. No lost material. No inconclusive results.
Say goodbye to hyperwelling with Countable PCR
If you’re still hyperwelling, you’re spending more time and resources to see less. Countable PCR puts the full sample to work — so you get rare event detection that’s more sensitive, more scalable, and way less frustrating.


.jpg)