Introduction
Accurate and reproducible quantification of recombinant adeno-associated virus (AAV) vector titers is crucial in gene therapy research and manufacturing. While digital PCR (dPCR) is employed as a quantitative titering method, its reliability is compromised by technical challenges. Specifically, inconsistencies in partitioning volume within dPCR systems affect quantitation accuracy. Furthermore, the limited dynamic counting range of these systems necessitates precise dilutions of viral vector samples to fall within this range, potentially leading to counting errors and repeated experiments [1].
In addition to the limitations of dPCR, challenges also arise from the specific genomic elements targeted using PCR-based titering. Due to the ubiquitous inclusion in the design of AAV vectors, the ITR2 region is often used for titering by PCR-based methods. However, the secondary structure of the ITRs often reduces PCR amplification efficiency and generates inconsistent results [2]. Mutations or deletions in the ITR further compromise titering accuracy. Additionally, ITR-based titering does not differentiate between functional and non-functional virions, leading to potential potency miscalculations [3]. The most direct way to confirm that an AAV stock contains its cargo and to determine its titer is to target the transgene, or gene of interest (GOI). However, because a new TaqMan assay must be developed and optimized for every new GOI, this approach is much less practical, inefficient, and costly.
Countable PCR directly addresses these limitations of traditional titering approaches. Unlike conventional dPCR, Countable PCR partitions samples into approximately 30 million spatially separated compartments, enabling single-molecule resolution across an exceptionally broad dynamic range—up to one million molecules. This eliminates the need for precise serial dilutions, reducing variability and simplifying workflows while maintaining high quantitation accuracy.
Countable PCR also overcomes the practical barriers of trasngene-specific titering with its support for Universal Multiplexing (UM) chemistry. UM uses unmodified, cost-effective primers and a universal probe to streamline assay design, allowing researchers to develop flexible and scalable assays for any transgene without the cost and complexity of custom TaqMan probe development. Together, these capabilities make Countable PCR a highly accurate, efficient, and practical solution for reliable AAV titer quantification.
This application note demonstrates the fundamentals of designing GOI-specific Countable PCR assays using TaqMan and UM chemistries for titer determination. It also presents the results of implementing these titering assays on AAV samples, demonstrating the following:
- The 6-log single molecule counting dynamic range of Countable PCR reduces the number of dilutions required for AAV titering, thereby minimizing variability.
- Countable Lab's UM chemistry offers flexibility in designing Gene of Interest (GOI)-specific assays with rapid turnaround times, saving both time and money.
Materials and methods
Design of GOI-specific assay using TaqMan hydrolysis probe chemistry
A TaqMan assay targeting the GOI, consisting of forward and reverse primers, and gene specific hydrolysis probe, was designed following best practices of oligo design. Additionally, a probe additive, an oligo partially complementary to the probe, was included to further quench the background fluorescence of the TaqMan probe, per guidelines outlined in the Countable PCR Reaction Preparation Guide.
Design of GOI-specific assay using Universal Multiplexing (UM) chemistry
The UM design was based on the forward and reverse primer pairs of the existing GOI-specific TaqMan assay, with only a UM adapter sequence added to the forward primer, with no additional purification.
AAV vector extraction
A small sample from a high-concentration AAV stock was extracted using standard protocol [4]. In brief, the sample was treated with DNase I (50 units) at 37°C for 10 minutes, followed by heat inactivation at 72°C for 10 minutes. Proteinase K (0.4 units) was added to the sample and incubated at 50°C for 60 minutes, followed by heat inactivation at 95°C for 20 minutes.
Sample dilution
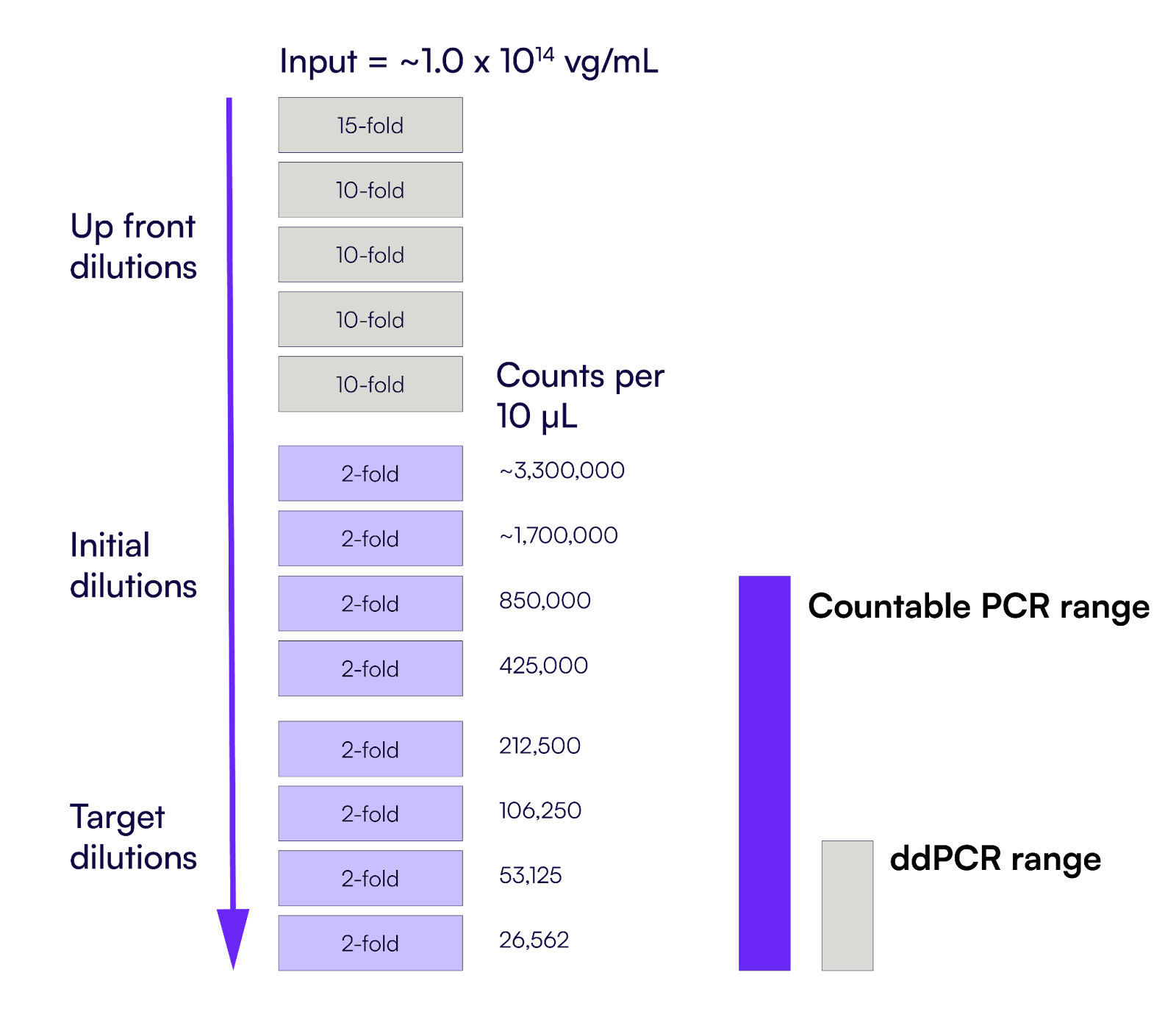
After AAV extraction, an aliquot of that material was diluted 15-fold, followed by four additional 10-fold dilution steps, by taking 5 µL of sample into 45 µL of buffer at each step (Figure 1). This “upfront dilution” scheme was needed for any AAV titering method that starts from a high concentration stock to reduce the test stock by 150,000-fold. From here, a series of 2-fold dilutions was done to bring the test stock concentration within the dynamic range of an assay.
Countable PCR workflow
Consumable materials needed for Countable PCR and the workflow are described in the Countable PCR Reaction Preparation User Guide. UM assays were run with the Countable Universal Multiplex Set A (KT0005).
Back calculation from molecule counts to AAV titer as vg/mL
To determine AAV titer, the counts obtained for a given dilution from the Countable PCR report were multiplied by the cumulative dilution factor, starting from the original stock. Therefore, the stock titer in viral genomes per mL (vg/mL) was calculated as:
Viral genomes per mL (vg/mL) = (Cumulative dilution factor * Counts at a specific dilution) / volume of extracted AAV sample loaded into assay in µL * 1000 μL/mL
This calculation was performed for at least three consecutive 10-fold dilution steps. The average of those titers was reported as the actual vector titer as vg/mL for that AAV batch.
Results
Broad dynamic range of Countable PCR reduces the requirement for extensive sample dilution
Typical AAV sample input is ~1.0x1014 vg/mL. Because dPCR has a narrow dynamic range for quantification, to get the most accurate Poisson-based correction, vector stocks are recommended to be diluted to a concentration <1,000 copies per reaction to guarantee single-molecule resolution [5, 6]. This means as much as a 1x109 fold dilution over 13 pipetting events (Figure 1). Incorrect dilution schemes often exceed the dynamic range of the dPCR systems, leading to titer calculation inaccuracy and necessitating the need to repeat measurements. Minor pipetting errors introduced at each dilution event also exacerbate overall measurement error, adding variability in titering. The requirement for extensive dilution reinforces the need for optimized pre-assay dilutions or a method more forgiving of dilution errors [7].
In comparison, Countable PCR has a single-molecule dynamic range of a million, requiring much fewer dilutions for titer determination. Fewer dilutions mean less work and fewer measurement errors, reducing overall measurement variability. Furthermore, with a wider window to fall within, there is less concern about over- or under- diluting a sample to hit the quantitative range.
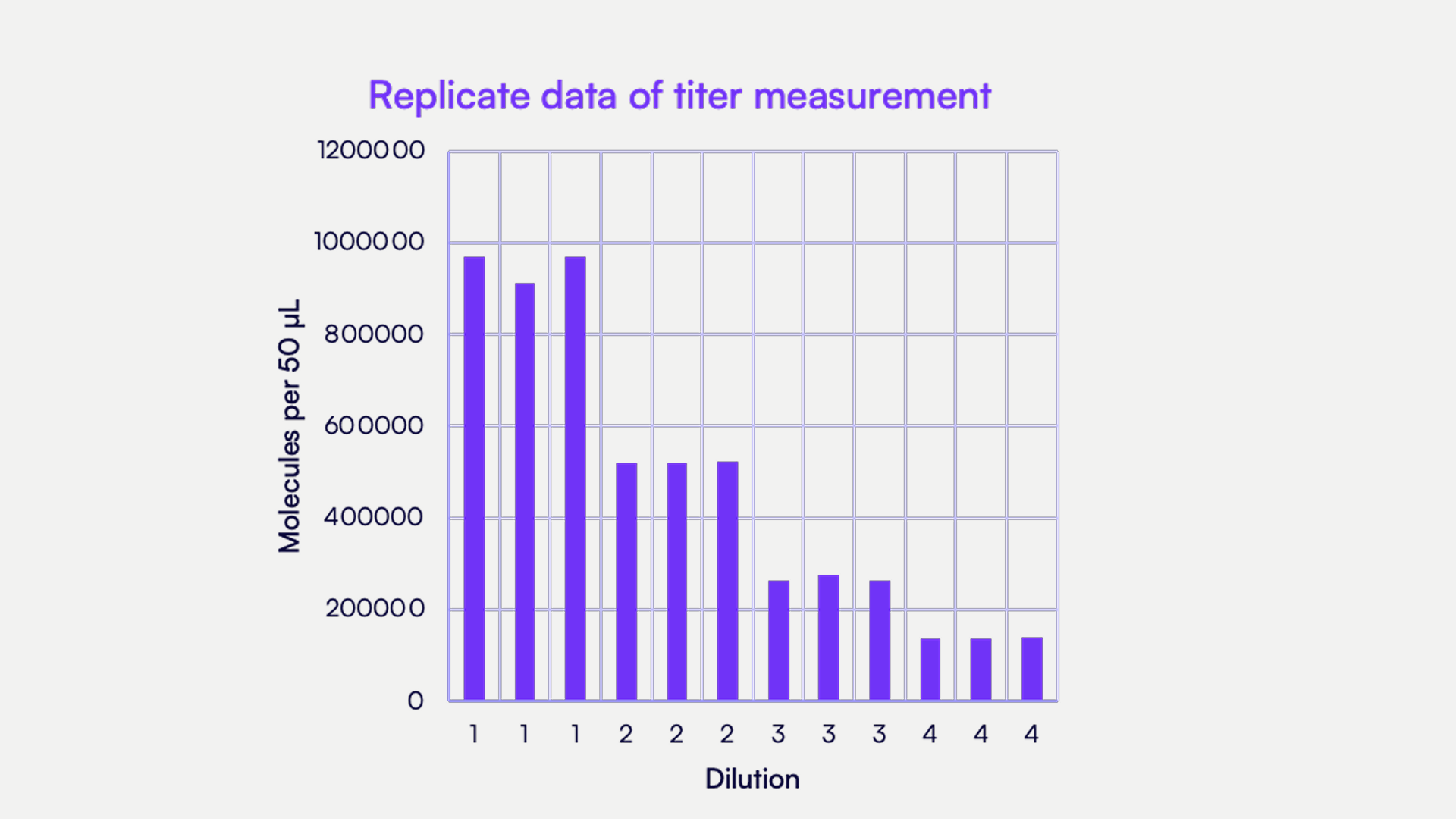
Countable PCR demonstrates robust performance across different batches
Using the TaqMan designs to evaluate the performance of Countable PCR in reporting AAV titer, we conducted a titering experiment using an AAV sample prepared as three separate batches. For linearity evaluation, each batch sample was serially diluted over multiple 2-fold steps, down to a concentration that achieved target counts of about 200k, 100k, and 50k. Each dilution was run in technical triplicates. For each batch, across the three dilution points, the back-calculated titer values were highly consistent, demonstrating robust linearity of the Countable PCR assay.
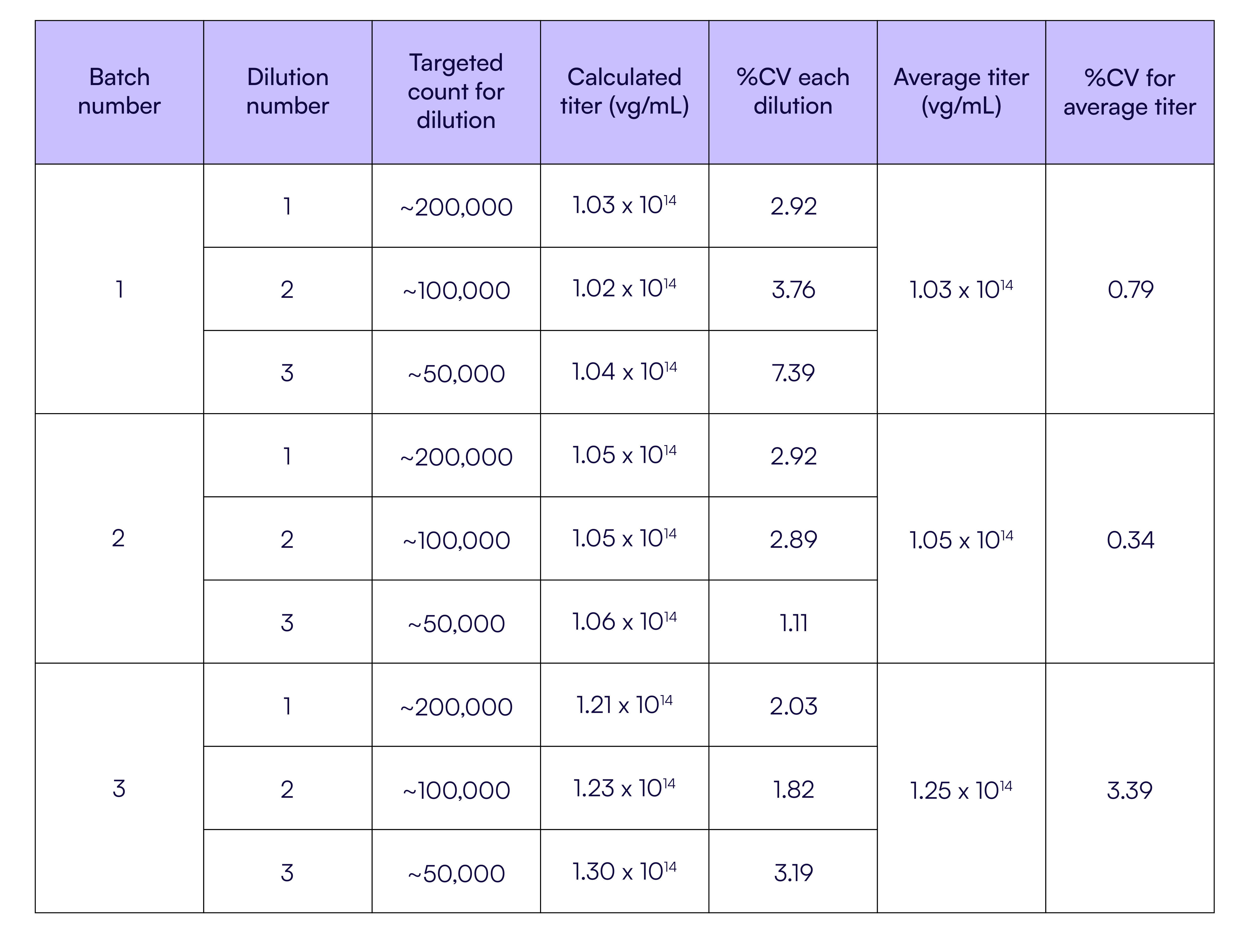
As shown in Figure 2, across three independent but controlled AAV manufacturing events, Countable PCR accurately called the titer of three batches that were expected to have the same yield, demonstrating excellent agreement with titers of 1.03 × 1014 vg/mL, 1.05 × 1014 vg/mL, and 1.25 × 1014 vg/mL for batches 1, 2, and 3, respectively. At the most dilute condition (~50k counts, dilution #3), the coefficient of variation (%CV) was 2.9%, 2.9%, and 2.0% for all three batches, respectively. The consistency of technical replicates within a batch supports the idea that this assay accurately compares titers between batches, and based on the observed %CV being below 1% for dilutions #1 and #2, it was sensitive enough to detect changes between production batches. The determined titers matched closely to an orthogonal analysis done by ddPCR that reported a titer of 7.88 × 1013 vg/mL for batch #1. Exact results are reported in Table 1.
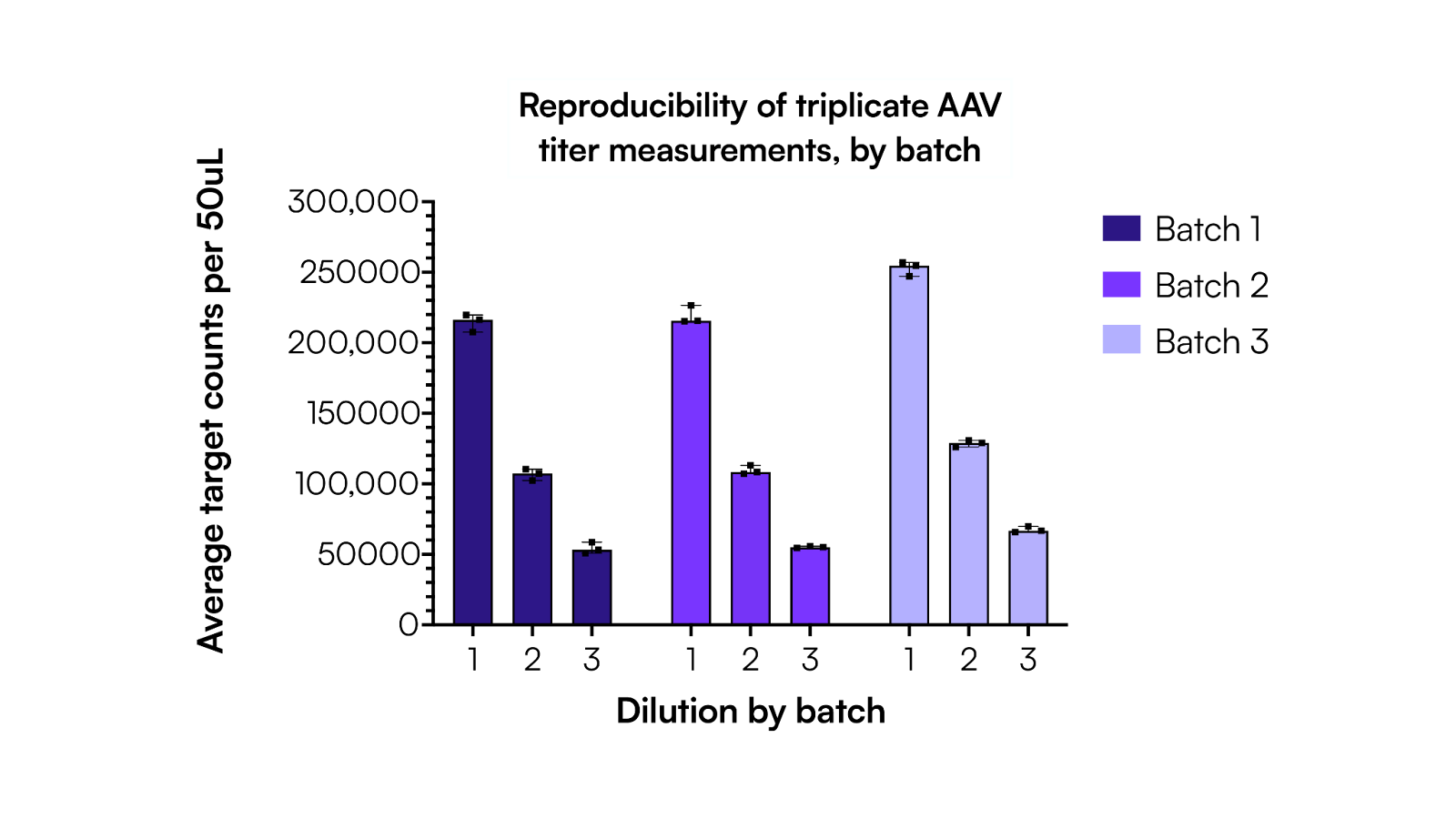
Table 1. Individual titer calculations for each dilution within a batch, including %CV batch that shows tight consistency, with overall the same calculated average titer when compared within the three dilutions by batch.
Universal Multiplexing (UM) chemistry is a low-cost but equally performing alternative to TaqMan chemistry for GOI-specific assay
While using an assay specifically against the GOI is more robust for AAV titering than using an assay of the ITR region, each new GOI requires developing a new GOI-specific assay. Therefore, though Countable PCR supports TaqMan probes, their use is cost-prohibitive and time-consuming.
To show Universal Multiplexing provides comparable performance, we demonstrated the interchangeability of a TaqMan assay with UM chemistry for AAV titering by evaluating the performance of a UM assay designed to target the same region on the AAV transgene as a TaqMan assay. To design a UM assay, the TaqMan probe was omitted, and the forward primer was modified to carry a UM adapter on the 5’ end. From design to testing, the process for UM assay validation took 3 days, as opposed to weeks for a typical TaqMan assay. Because only standard desalted primers are required, the cost of a UM assay is about only ~1/10th of that of a TaqMan assay, which requires a custom probe.
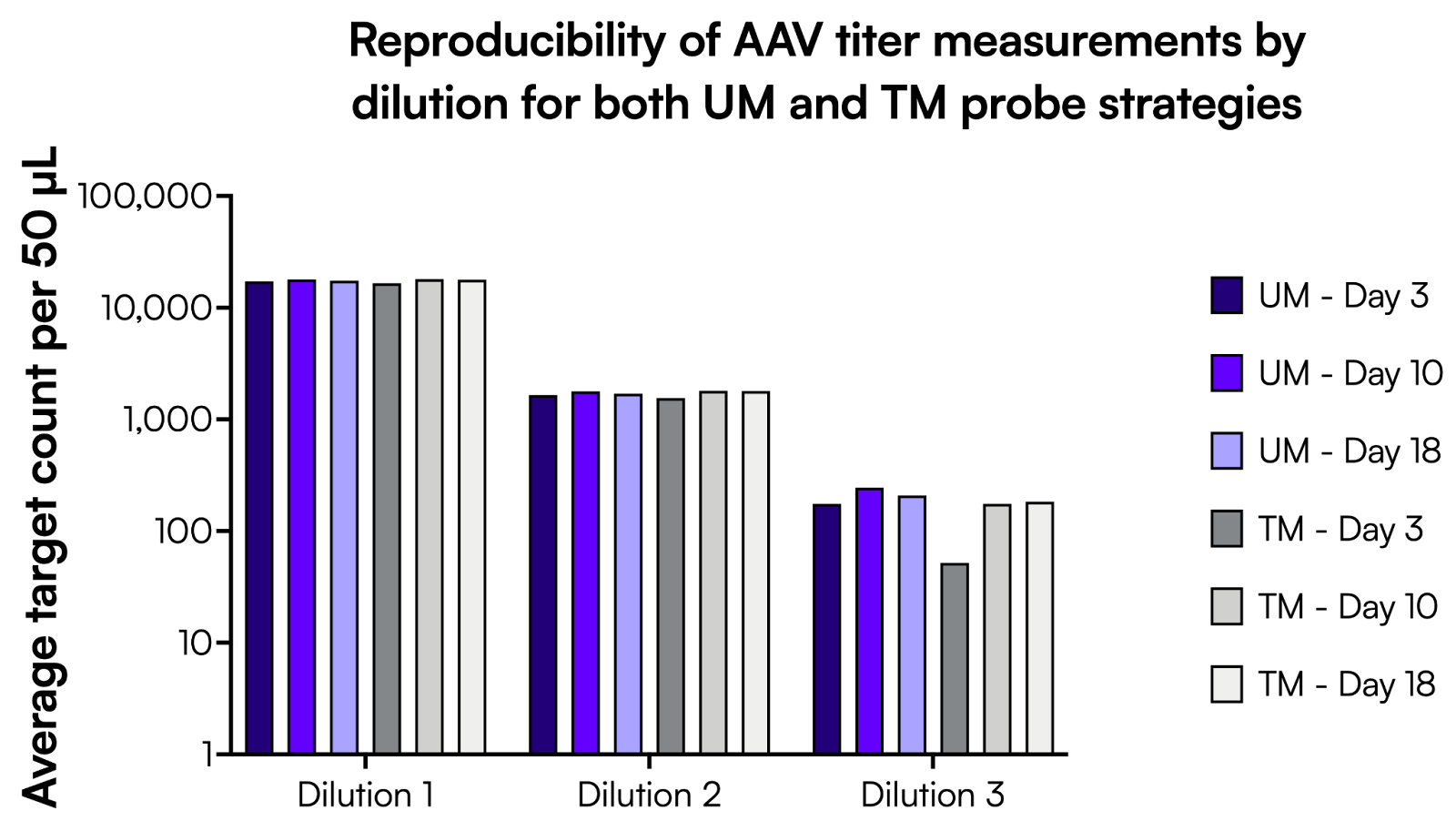
To show sensitivity and reproducibility, the AAV stock from manufacturing batch #1 was tested with the TaqMan assay and its UM equivalent, at three independent time points: 3, 10, and 18 days post-storage at -80°C. The sample was serially diluted in 10-fold steps to reach counts from 20,000 down to ~200 counts per 50 µL reaction, and run as technical triplicates for each dilution. The measured counts per 50 µL are shown in Figure 3. Overall, data show that both chemistries perform equally well on the Countable system
Conclusion
Countable PCR for AAV titer determination using TaqMan or UM chemistries showed excellent dilution linearity, as well as reproducibility between replicates and across batch manufacturing events.
The broad dynamic range of Countable PCR makes it ideal for precision applications like AAV titering, since fewer dilutions are required. Additionally, UM chemistry enables flexibility and cost savings for designing and optimizing assays using the Gene of Interest as the target. Collectively, these features reduce variability, save time and costs, and generate trustworthy, actionable data with less effort than any other system.
References
- <span id="resource-1"></span>Zanker, J. et al. Insight and Development of Advanced Recombinant Adeno-Associated Virus Analysis Tools Exploiting Single-Particle Quantification by Multidimensional Droplet Digital PCR. Human Gen. Ther. doi: /10.1089/hum.2021.182 Volume 33: No 17-18. 16 Sept 2022.
- <span id="resource-2"></span>Wang F, et al. A reliable and feasible qPCR strategy for titrating AAV vectors. Med Sci Monit Basic Res. 2013 Jul 5;19:187-93. doi: 10.12659/MSMBR.883968. PMID: 23828206; PMCID: PMC3706409.
- <span id="resource-3"></span>François, Achille et al. Accurate Titration of Infectious AAV Particles Requires Measurement of Biologically Active Vector Genomes and Suitable Controls. Molecular Therapy Methods & Clinical Development, Volume 10, 223 - 236
- <span id="resource-4"></span>Werling NJ, Satkunanathan S, Thorpe R, Zhao Y. Systematic Comparison and Validation of Quantitative Real-Time PCR Methods for the Quantitation of Adeno-Associated Viral Products. Hum Gene Ther Methods. 2015 Jun;26(3):82-92. doi: 10.1089/hgtb.2015.013. Epub 2015 Jun 9. PMID: 25953194; PMCID: PMC4492554.
- <span id="resource-5"></span>Sanmiguel J, Gao G, Vandenberghe LH. Quantitative and Digital Droplet-Based AAV Genome Titration. Methods Mol Biol. 2019;1950:51-83. doi: 10.1007/978-1-4939-9139-6_4. PMID: 30783968; PMCID: PMC7377043.
- <span id="resource-6"></span>Suoranta T, Laham-Karam N, Ylä-Herttuala S. Optimized Protocol for Accurate Titration of Adeno-Associated Virus Vectors. Hum Gene Ther. 2021 Oct;32(19-20):1270-1279. doi: 10.1089/hum.2020.318. Epub 2021 May 6. PMID: 33560161.
- <span id="resource-7"></span>Lengler J, Gavrila M, Brandis J, Palavra K, Dieringer F, Unterthurner S, Fuchsberger F, Kraus B, Bort JAH. Crucial aspects for maintaining rAAV stability. Sci Rep. 2024 Nov 12;14(1):27685. doi: 10.1038/s41598-024-79369-0. PMID: 39533000; PMCID: PMC11557909.
Introduction
Accurate and reproducible quantification of recombinant adeno-associated virus (AAV) vector titers is crucial in gene therapy research and manufacturing. While digital PCR (dPCR) is employed as a quantitative titering method, its reliability is compromised by technical challenges. Specifically, inconsistencies in partitioning volume within dPCR systems affect quantitation accuracy. Furthermore, the limited dynamic counting range of these systems necessitates precise dilutions of viral vector samples to fall within this range, potentially leading to counting errors and repeated experiments [1].
In addition to the limitations of dPCR, challenges also arise from the specific genomic elements targeted using PCR-based titering. Due to the ubiquitous inclusion in the design of AAV vectors, the ITR2 region is often used for titering by PCR-based methods. However, the secondary structure of the ITRs often reduces PCR amplification efficiency and generates inconsistent results [2]. Mutations or deletions in the ITR further compromise titering accuracy. Additionally, ITR-based titering does not differentiate between functional and non-functional virions, leading to potential potency miscalculations [3]. The most direct way to confirm that an AAV stock contains its cargo and to determine its titer is to target the transgene, or gene of interest (GOI). However, because a new TaqMan assay must be developed and optimized for every new GOI, this approach is much less practical, inefficient, and costly.
Countable PCR directly addresses these limitations of traditional titering approaches. Unlike conventional dPCR, Countable PCR partitions samples into approximately 30 million spatially separated compartments, enabling single-molecule resolution across an exceptionally broad dynamic range—up to one million molecules. This eliminates the need for precise serial dilutions, reducing variability and simplifying workflows while maintaining high quantitation accuracy.
Countable PCR also overcomes the practical barriers of trasngene-specific titering with its support for Universal Multiplexing (UM) chemistry. UM uses unmodified, cost-effective primers and a universal probe to streamline assay design, allowing researchers to develop flexible and scalable assays for any transgene without the cost and complexity of custom TaqMan probe development. Together, these capabilities make Countable PCR a highly accurate, efficient, and practical solution for reliable AAV titer quantification.
This application note demonstrates the fundamentals of designing GOI-specific Countable PCR assays using TaqMan and UM chemistries for titer determination. It also presents the results of implementing these titering assays on AAV samples, demonstrating the following:
- The 6-log single molecule counting dynamic range of Countable PCR reduces the number of dilutions required for AAV titering, thereby minimizing variability.
- Countable Lab's UM chemistry offers flexibility in designing Gene of Interest (GOI)-specific assays with rapid turnaround times, saving both time and money.
Materials and methods
Design of GOI-specific assay using TaqMan hydrolysis probe chemistry
A TaqMan assay targeting the GOI, consisting of forward and reverse primers, and gene specific hydrolysis probe, was designed following best practices of oligo design. Additionally, a probe additive, an oligo partially complementary to the probe, was included to further quench the background fluorescence of the TaqMan probe, per guidelines outlined in the Countable PCR Reaction Preparation Guide.
Design of GOI-specific assay using Universal Multiplexing (UM) chemistry
The UM design was based on the forward and reverse primer pairs of the existing GOI-specific TaqMan assay, with only a UM adapter sequence added to the forward primer, with no additional purification.
AAV vector extraction
A small sample from a high-concentration AAV stock was extracted using standard protocol [4]. In brief, the sample was treated with DNase I (50 units) at 37°C for 10 minutes, followed by heat inactivation at 72°C for 10 minutes. Proteinase K (0.4 units) was added to the sample and incubated at 50°C for 60 minutes, followed by heat inactivation at 95°C for 20 minutes.
Sample dilution
After AAV extraction, an aliquot of that material was diluted 15-fold, followed by four additional 10-fold dilution steps, by taking 5 µL of sample into 45 µL of buffer at each step (Figure 1). This “upfront dilution” scheme was needed for any AAV titering method that starts from a high concentration stock to reduce the test stock by 150,000-fold. From here, a series of 2-fold dilutions was done to bring the test stock concentration within the dynamic range of an assay.
Countable PCR workflow
Consumable materials needed for Countable PCR and the workflow are described in the Countable PCR Reaction Preparation User Guide. UM assays were run with the Countable Universal Multiplex Set A (KT0005).
Back calculation from molecule counts to AAV titer as vg/mL
To determine AAV titer, the counts obtained for a given dilution from the Countable PCR report were multiplied by the cumulative dilution factor, starting from the original stock. Therefore, the stock titer in viral genomes per mL (vg/mL) was calculated as:
Viral genomes per mL (vg/mL) = (Cumulative dilution factor * Counts at a specific dilution) / volume of extracted AAV sample loaded into assay in µL * 1000 μL/mL
This calculation was performed for at least three consecutive 10-fold dilution steps. The average of those titers was reported as the actual vector titer as vg/mL for that AAV batch.
Results
Broad dynamic range of Countable PCR reduces the requirement for extensive sample dilution
Typical AAV sample input is ~1.0x1014 vg/mL. Because dPCR has a narrow dynamic range for quantification, to get the most accurate Poisson-based correction, vector stocks are recommended to be diluted to a concentration <1,000 copies per reaction to guarantee single-molecule resolution [5, 6]. This means as much as a 1x109 fold dilution over 13 pipetting events (Figure 1). Incorrect dilution schemes often exceed the dynamic range of the dPCR systems, leading to titer calculation inaccuracy and necessitating the need to repeat measurements. Minor pipetting errors introduced at each dilution event also exacerbate overall measurement error, adding variability in titering. The requirement for extensive dilution reinforces the need for optimized pre-assay dilutions or a method more forgiving of dilution errors [7].
In comparison, Countable PCR has a single-molecule dynamic range of a million, requiring much fewer dilutions for titer determination. Fewer dilutions mean less work and fewer measurement errors, reducing overall measurement variability. Furthermore, with a wider window to fall within, there is less concern about over- or under- diluting a sample to hit the quantitative range.

Countable PCR demonstrates robust performance across different batches
Using the TaqMan designs to evaluate the performance of Countable PCR in reporting AAV titer, we conducted a titering experiment using an AAV sample prepared as three separate batches. For linearity evaluation, each batch sample was serially diluted over multiple 2-fold steps, down to a concentration that achieved target counts of about 200k, 100k, and 50k. Each dilution was run in technical triplicates. For each batch, across the three dilution points, the back-calculated titer values were highly consistent, demonstrating robust linearity of the Countable PCR assay.
As shown in Figure 2, across three independent but controlled AAV manufacturing events, Countable PCR accurately called the titer of three batches that were expected to have the same yield, demonstrating excellent agreement with titers of 1.03 × 1014 vg/mL, 1.05 × 1014 vg/mL, and 1.25 × 1014 vg/mL for batches 1, 2, and 3, respectively. At the most dilute condition (~50k counts, dilution #3), the coefficient of variation (%CV) was 2.9%, 2.9%, and 2.0% for all three batches, respectively. The consistency of technical replicates within a batch supports the idea that this assay accurately compares titers between batches, and based on the observed %CV being below 1% for dilutions #1 and #2, it was sensitive enough to detect changes between production batches. The determined titers matched closely to an orthogonal analysis done by ddPCR that reported a titer of 7.88 × 1013 vg/mL for batch #1. Exact results are reported in Table 1.

Table 1. Individual titer calculations for each dilution within a batch, including %CV batch that shows tight consistency, with overall the same calculated average titer when compared within the three dilutions by batch.

Universal Multiplexing (UM) chemistry is a low-cost but equally performing alternative to TaqMan chemistry for GOI-specific assay
While using an assay specifically against the GOI is more robust for AAV titering than using an assay of the ITR region, each new GOI requires developing a new GOI-specific assay. Therefore, though Countable PCR supports TaqMan probes, their use is cost-prohibitive and time-consuming.
To show Universal Multiplexing provides comparable performance, we demonstrated the interchangeability of a TaqMan assay with UM chemistry for AAV titering by evaluating the performance of a UM assay designed to target the same region on the AAV transgene as a TaqMan assay. To design a UM assay, the TaqMan probe was omitted, and the forward primer was modified to carry a UM adapter on the 5’ end. From design to testing, the process for UM assay validation took 3 days, as opposed to weeks for a typical TaqMan assay. Because only standard desalted primers are required, the cost of a UM assay is about only ~1/10th of that of a TaqMan assay, which requires a custom probe.
To show sensitivity and reproducibility, the AAV stock from manufacturing batch #1 was tested with the TaqMan assay and its UM equivalent, at three independent time points: 3, 10, and 18 days post-storage at -80°C. The sample was serially diluted in 10-fold steps to reach counts from 20,000 down to ~200 counts per 50 µL reaction, and run as technical triplicates for each dilution. The measured counts per 50 µL are shown in Figure 3. Overall, data show that both chemistries perform equally well on the Countable system

Conclusion
Countable PCR for AAV titer determination using TaqMan or UM chemistries showed excellent dilution linearity, as well as reproducibility between replicates and across batch manufacturing events.
The broad dynamic range of Countable PCR makes it ideal for precision applications like AAV titering, since fewer dilutions are required. Additionally, UM chemistry enables flexibility and cost savings for designing and optimizing assays using the Gene of Interest as the target. Collectively, these features reduce variability, save time and costs, and generate trustworthy, actionable data with less effort than any other system.
References
- <span id="resource-1"></span>Zanker, J. et al. Insight and Development of Advanced Recombinant Adeno-Associated Virus Analysis Tools Exploiting Single-Particle Quantification by Multidimensional Droplet Digital PCR. Human Gen. Ther. doi: /10.1089/hum.2021.182 Volume 33: No 17-18. 16 Sept 2022.
- <span id="resource-2"></span>Wang F, et al. A reliable and feasible qPCR strategy for titrating AAV vectors. Med Sci Monit Basic Res. 2013 Jul 5;19:187-93. doi: 10.12659/MSMBR.883968. PMID: 23828206; PMCID: PMC3706409.
- <span id="resource-3"></span>François, Achille et al. Accurate Titration of Infectious AAV Particles Requires Measurement of Biologically Active Vector Genomes and Suitable Controls. Molecular Therapy Methods & Clinical Development, Volume 10, 223 - 236
- <span id="resource-4"></span>Werling NJ, Satkunanathan S, Thorpe R, Zhao Y. Systematic Comparison and Validation of Quantitative Real-Time PCR Methods for the Quantitation of Adeno-Associated Viral Products. Hum Gene Ther Methods. 2015 Jun;26(3):82-92. doi: 10.1089/hgtb.2015.013. Epub 2015 Jun 9. PMID: 25953194; PMCID: PMC4492554.
- <span id="resource-5"></span>Sanmiguel J, Gao G, Vandenberghe LH. Quantitative and Digital Droplet-Based AAV Genome Titration. Methods Mol Biol. 2019;1950:51-83. doi: 10.1007/978-1-4939-9139-6_4. PMID: 30783968; PMCID: PMC7377043.
- <span id="resource-6"></span>Suoranta T, Laham-Karam N, Ylä-Herttuala S. Optimized Protocol for Accurate Titration of Adeno-Associated Virus Vectors. Hum Gene Ther. 2021 Oct;32(19-20):1270-1279. doi: 10.1089/hum.2020.318. Epub 2021 May 6. PMID: 33560161.
- <span id="resource-7"></span>Lengler J, Gavrila M, Brandis J, Palavra K, Dieringer F, Unterthurner S, Fuchsberger F, Kraus B, Bort JAH. Crucial aspects for maintaining rAAV stability. Sci Rep. 2024 Nov 12;14(1):27685. doi: 10.1038/s41598-024-79369-0. PMID: 39533000; PMCID: PMC11557909.